Original Article
, Volume: 13( 2)Zinc(II) Complexes Derived from N,N-Donors
- *Correspondence:
- Sahar I. MostafaDepartment of Chemistry, Faculty of Science, Mansoura University, Egypt, Tel: 002-01008502625; Fax: 0020502246781; E-Mail: sihmostafa@gmail.com
Received: June 06, 2018; Accepted: July 10, 2018; Published: July 16, 2018
Citation: Annan NA, Butler IS, Yahia El-Lazeik, Mostafa SI. Zinc(II) Complexes Derived from N,N-Donors. Inorg Chem Ind J. 2018;13(2):126.
Abstract
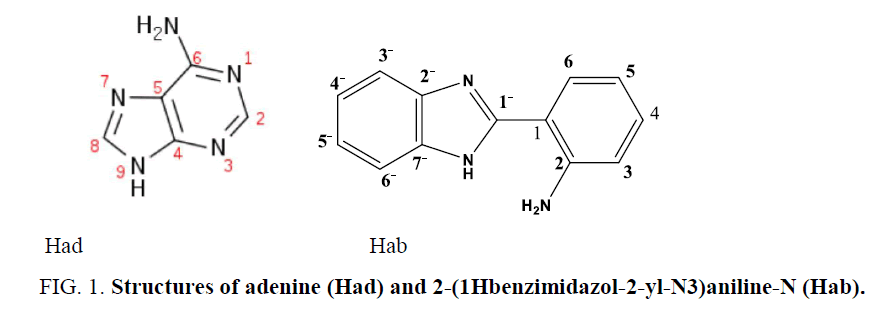
Synthesis and characterization of zinc(II) complexes, [Zn(ad)Cl(H2O)] and [Zn(Hab)2](ClO4)2 {Had = adenine, Hab = 2-(1Hbenzimidazol-2-yl-N3)aniline-N} are reported. The ligands coordinate Zn(II) ion via two N-centers. Zn(II) atom is coordinating in tetrahedral structure, cyclic N7 after deprotonation of N9, NH2 (ad-), chloride and water molecule, and imidazole N and NH2 from two Hab ligands, respectively.Keywords
Adenine, Complexes, Spectra, Mass, Zinc
Introduction
Benzimidazoles (heterocyclic aromatic compound consists of a benzene ring fused with imidazole) [1], are involved in variety of biological processes [2-5]. They are of interest, since reported that the substitution at 1, 2 and 5 positions is very important for pharmacological actions [6]. Benzimidazole scaffold is considered as a useful structural motif for the development of therapeutic agents that display a broad spectrum of pharmacological activities. Many DNA minor groove binders containing one or more imidazole show effective anticancer and antiparasitic activities [7]. Bis-benzimidazole derivatives Zn(II) [7], Co(II) [7], Cu(II) [8] complexes have been reported. The monomeric and polymeric benzimidazole and malonic acid mixed-ligands complexes of Co (II) and Cu (II) have been prepared and their structures discussed [9]. The X-ray crystal structure of [Cr2 (2gb)4(µ-OH)2](ClO4)4.5H2O (2gb = 2-Guanidinobenzimidazole) showed the presence of two 2gb moieties and a symmetric (µ-OH)2 bridge [10].
The 2-(2-aminophenyl)-1H-benzimidazole (Hab) may behave as a bidentate ligand through its aniline group assisted by the imidazole nitrogen. In the free ligand a strong H-bond indicates the natural site for metal ion coordination [11]. This feature is also confirmed by the X-ray crystal structure of [Ni(Hab)2(H2O)2](NO3)2, [Ag(Hab2]NO3, [Zn(Hab)Cl2] and [Zn(Hab)2(H2O)](NO3)2 [11-14].
Adenine is one of the two purine nucleobases (the other being guanine) used in forming nucleotides of the nucleic acids. In DNA, adenine (A) binds to thymine (T) by two H-bonds to assist stabilizing nucleic acid structures, while in RNA, adenine (A) binds to uracil (U). Adenine was early prepared from polymerization of ammonia with hydrogen cyanide [15]. Adenine complexes with Mn(II), Fe(II), Co(II), Ni(II) and Cu(II) ions have been reported [16,17]. In addition, the octahedral complexes [M(CA)2(Had)X2] {M(II) = Co, Ni, Cu, Zn, CA = caffeine, Had = adenine, X = SCN} have been synthesized and investigated [18]. The X-ray structure of [Cu(acac)2(Had)].EtOH complex reveals a square-pyramidal geometry, with acetylacetone occupies the equatorial positions while adenine is in the axial position and is coordinated through the N7 atom [19].
Moreover, linear chainlike polymeric complexes of Had with the first row transition elements have been isolated [20]. The Xray crystal structure of the complex [(CH3Hg)2(ad)].CH3CH2OH has been established [21]. Mixed Pt(II) adenine derivative complexes, [Pt(L)2(ox)], [Pt(ad?)2(ox)] (ox = oxalate, L = 2-chloro-N6-(benzyl)-9- isopropyladenine derivatives) have been reported and the X-ray structure of [Pt(2,4-diOMeL)2(ox)].2DMF proved slight distorted square?planar geometry, coordinates through the N7 atom of adenine moieties [22].
It is interesting to turn our attention to investigate the reaction of bifunctional N,N-donors (FIG. 1) , 2-(1Hbenzimidazol-2-yljN3) aniline-N (Hab) and adenine (Had) with the biocompatible Zn(II) ion, which can be utilized to minimize the toxicity, as it is familiar with the biological environment of the human body.
Method
All reagents and solvents were purchased from Alfa/Aesar and all manipulations were performed under aerobic conditions using materials and solvents as received. DMSO-d6 was used for the NMR measurements referenced against TMS.
Measurements
Elemental analyses (C, H, N) were performed in the Microanalytical Unit, Department of Chemistry, Cairo University. Zinc contents were determined by complexometric titration. Infrared spectra were recorded on a Nicolet 6700 Diamond ATR spectrometer in the 4000- 200 cm-1 range. NMR spectra were measured on Varian Mercury 200-, 300- and 500- MHz spectrometer, in DMSO d6 using TMS as reference. Mass spectra (ESI-MS) were recorded using LCQ Duo and double focusing MS25RFA instruments, respectively. Electronic spectra were recorded in DMF using a Hewlett-Packard 8453 spectrophotometer. Thermal analysis measurements were made in the 20? 1000o C range at a heating rate of 20o C min-1 using Ni and NiCo as references, on a on a TA instrument TGA model Q500Analyzer TGA-50. Molar conductivity measurements were carried out at room temperature on a YSI Model 32 conductivity bridge.
Preparation of Complex
1. [Zn(ad)Cl(H2O)].H2O
Adenine, Had (0.033 g, 0.25 mmol) in MeOH containing KOH (0.014 g, 0.25 mmol; 5 mL) was added to an aqueous solution of ZnCl2 (0.034 g, 0.25 mmol; 5 ml). The reaction mixture was heated under reflux for 4h. The pale yellow precipitate was filtered off, washed with water, MeOH and dried in vacuo. Yield: 68%. Elemental Anal.: C, 61.8; H, 4.7; N, 15.7 (C46H42N10O2P2Zn); Found: C, 61.9; H, 4.7; N, 15.6% (C5ClH8N5O2Zn) Calcd. C, 22.14; H, 2.95; N, 25.83; Cl, 13.10; Zn, 24.17 Found: C, 22.20; H, 3.01; N, 25.85; Cl, 13.15; Zn, 24.02%. Conductivity data (10-3 M in DMF): M = 3.11 ohm-1 cm2 mol-1.
2. [Zn(Hab)2](ClO4)2.2H2O
An aqueous solution of Zn(ClO4)2 (0.132 g, 0.5 mmol; 5 mL) was added to Hba (0.105 g, 0.5 mmol) in EtOH (15 mL). The reaction mixture was stirred and heated under reflux for 3h. The yellow solid was filtered off, washed with water, EtOH and air-dried. Yield: 72%. Elemental Anal.: (C26Cl2H26N6O10Zn) C, 43.42; Cl, 9.88; H, 3.62; N, 11.69; Zn, 9.12; Found: C, 43.40; Cl, 9.78; H, 3.54; N, 11.60; Zn, 9.03%. Conductivity data (10-3 M in DMF): M = 189.06 ohm-1 cm2 mol-1.
Results and Discussion
The elemental analyses for the complexes are in agreement with the assigned formulae. The molar conductivities (?M) in DMF at room temperature suggest 1:2 electrolytic nature of complexes (2), while, complex (1) shows non-electrolytic nature [23,24].
Vibrational spectra
The comparison of the IR spectra of Had with those of the complexes well explain and prove the modes of chelation of Had. Földesi et al. [25], reported that on deuteration of Had, the C(8)-H proton changes to C(8)-D (in D2O) > 100ºC while C(2)-H does not exchange. Furthermore, at Room temperature and up to 80oC, N(9)-D and ND2 were deuterated, while H(2)?D and N(9)-D do not exchange [26]. This feature may be attributed to the complicated catalytic conditions required for adenine H?D exchange reaction [27]. Had displays bands at 3370 and 3294 cm-1 arise from νas(NH2) and νs(NH2) stretches, respectively, while the strong bands at 3109 and 1330 cm-1 are assigned to ν(N9H) and d(N9H) stretches, respectively [17,28]. Band observed at 1665 cm-1 is due to δ(NH2) vibration, while those at 1600, 1503 and 1250 cm-1, are assigned to azomethine ν(C7=N8), ν(C6=N1) and ν(C2=N3) stretches, respectively [29-31]. The bands at 867 and 800 cm-1 are due to ?(NH2), while that at 1022 and 1005 cm-1 is attributed to ?(NH2) [17,32].
In complex (1), ?(NH) and d(NH) stretches are not observed, while νs(NH2), νas(NH2), ?(C1=N6), ?(C7=N8) and ?(C2=N3) stretches are shifted to lower wave number, indicating mononegative bidentate behaviour of ad-, coordinating Zn(II) ion through NH2 and (N7) nitrogen centers, after deprotonated of N9, forming five-membered chelate ring (TABLE 1) [32].
| Compound | νas(NH2) | νs(NH2) | ν(NH) | ν(C=N) & n(C-N) | ν(C6=N1) | ν(X2=N3) |
|---|---|---|---|---|---|---|
| Had | 3370 | 3294 | 3109 | 1600* | 1503 | 1250 |
| [Zn(ad)Cl(H2O)] (1) | 3443 | 3352 | 3107 | 1595* | 1500 | 1224 |
| Hab | 3379 | 3169 | 3056 | 1620 1320 | 1597 | |
| [Zn(Hab)2](ClO4)2 (2) | 3342 | 3179 | 3054 | 1589 1318 | 1569 |
TABLE 1. IR spectral data of Zn(II) complexes.
The IR spectrum of Hab shows medium bands at 3379, 3269 and 3056 cm-1 due to the νas(NH2), νs(NH2) and ν(NH) stretches, respectively [11]. The medium bands at 1620 and 1320 cm-1 in the free ligand is due to ν(C=N) and ν(C?N) stretches, respectively. In complex (2), the νas(NH2), νs(NH2) and ν(C=N) stretch observed at 3342, 31779 and 1589 cm-1 indicating the involvement of amino and azomethine-N atoms in coordination in neutral bidentate manner (TABLE 1), while the ν(NH) stretch is remaining more or less in the same position in the complex; i.e., plays no role in coordination [33,34]. The spectrum of (2) exhibits two bands at 1096 cm-1 (strong) and 690 cm-1 (medium) due to ν3(F2) and ν4(F2) of uncoordinated ClO4-, respectively [33,34].
NMR spectra
The 1H NMR spectrum of free Had exhibits sharp singlets at d 8.30, 8.25 and 7.38 ppm attributed to H2, H8 and NH2, respectively. The broad singlet d 13.00 ppm is due to N9-H proton. In complex (1), N9-H proton signal is missed indicating the coordination of Zn(II) to N7 after deprotonation of N9 [35], while the NH2 signal is shifted clearly upfield (d 7.00 ppm), indicating the complexation of ad- through the deprotonated N-7 and amino N-atoms in bidentate manner.
The 1H NMR spectrum of Hab shows sharp singlets at d 12.65, 7.24 ppm due to NH and NH2, respectively. A group of doublets and triplets in the range of d 6.63-7.84 ppm is attributed to the aromatic protons. In complex (2), a clear downfield shift in both NH2 and aromatic protons signals is observed indicating the coordination through amino and imidazole N-atoms in neutral bidentate behavior (TABLE 2).
| Compound | NH(H(1) S | NH2(H(2) S | (H(3,d ) | (H(4, t ) | (H(5, t ) | (H(6,d ) | (H(3-,d) |
|---|---|---|---|---|---|---|---|
| Had | 13 | 7.38 | 8.30* | 8.25** | |||
| [Zn(ad)Cl(H2O)] (1) | _ | 6.99 | 8.15* | 8.65** | |||
| Hab | 12.65 | 7.24 | 6.83 | 6.63 | 6.65 | 7.83 | 7.64 |
| [Zn(Hab)2](ClO4)2 (2) | 12.91 | 7.65 | 7.9 | 6.66 | 6.67 | 6.93 | 7.89 |
TABLE 2. 1H NMR spectral data of Zn(II) complexes.
Electronic spectra
The UV-Vis absorption spectra of Had, Hab, bpy and their Zn(II) complexes in DMSO were recorded at room temperature. The absorption spectra of the free ligands show band near 270 and 333 nm, which are due to ?????* transitions of phenyl ring and H-bond induced changes of amine NH (intraligand CT), respectively [36,37]. The spectra of the complexes show band in the 350- 370 nm range can be attributed to MLCT (to the ??* orbital of bpy or phenyl ring) [36,37].
Mass spectra
The mass spectral data of the complexes show molecular ion peaks are in agreement and confirmed their assigned formulae. The observed fragmentation patterns are corresponding to successive degradations of the molecule with stepwise ligand loss.
The spectrum of (1) shows peak at m/z 505.8 (Calcd. 056.0 with 12% abundance represents the molecular ion [Zn(ad)Cl]2+. The peaks at 252.6 and 198.1, are corresponding to [Zn(ad)Cl]+ (Calcd. 253.0) and [Zn(ad)]+ (Calcd. 199.5) fragments, respectively [38].
Complex (2) shows fragmentation pattern corresponding to successive degradation of Hab molecule. The signals at m/z 485.7 (Calcd. 483.5) and 275.3 (Calcd. 274.5) with 80% abundance represent the molecular ions, [Zn(ab)2]+ and [Zn(ab)]+ fragments, respectively [39,40].
Thermal studies
The thermogravimetric (TG) technique was applied to study the thermal stability and degradation behavior of the reported Zn(II) complexes. The observed mass-loss stages were arising from the elimination of hydrated water, coordinated water and/ or Cl2, and O2, followed by decomposition of the ligand units [34,36,39]. The weight loss observed below 130º C is due to dehydration; as the color changed to be deeper [33,34].
The decomposition of ad- or Hab was taken place via the break-down of the weak hetero (N-C) bonds [33,34,41].
The TGA of (1), [Zn(ad)(H2O)Cl].H2O, shows inflections in the ranges of 30?125, 126 ? 490 and 491 - 680º C, attributed to the loss of water of crystallization (Calcd. 6.6, Found 6.3%), coordinated H2O, ½ Cl2 and ½ N2 species (Calcd. 24.9, Found 25.5%) and C5H4N3 fragment (Calcd. 39.1, Found 38.7%), leaving residue of Zn(II) nitride (Calcd. 27.6, Found 30.1%) [42].
The thermogram of (2), [Zn(ab)2](ClO4)2.2H2O, shows the first step weight loss of 5.2% between 35 and 118º C, which corresponds to the release of two molecules of water of crystallization per molecule of complex and (Calcd. 5.0%) [42]. The second decomposition (119 - 327º C), is attributed to the loss of Cl2 and 4O2 (Calcd. 27.7, Found 27.5%), The third decomposition occurs between 328 and 500º C, arising from the elimination of two C6H5N fragments (Calcd. 25.3, Found 24.8%). The fourth weight loss (501 ? 650 ºC), is due to the loss of two C6H4 fragments (Calcd. 21.2, Found 21.9%), leaving residue of Zn(II) nitride with 14.7% [36].
Conclusion
Zn(II) complexes with multi-dentate N,N-donors {adenine (Had) and 2-(1Hbenzimidazol-2-yl-N3)aniline-N (Hab)} were preparation and their structures were discussed based on physico-chemical measurements. The complexation of Had and Hab, Zn(II) ion is four-coordinating tetrahedrals.
References
- Barker HA, Smyth RD, Weissbach H,et al. J Biol Chem, 1960;235:480-288.
- Pawar NS, Dalal DS and Shimpi SR. Eur J Pharm Sci, 2004;21:115-118.
- Hisano T, Ichikawa M and Tsumoto K. Chem Pharm Bull, 1982;30:2996-3004.
- Özden S, Atabey D, Yildiz S et al. Med Chem, 2005;13:1587-1497;
- Kühler TC, Swanson M, Christenson B, et al. J Med Chem, 2002;45:4282-4299.
- Carcanague D, Shue Y, Wuonola M, et al. J Med Chem, 2002; 45:4300-4309.
- Liu S, Cao W, Yu L, et al. Dalton Trans, 2013;42:5932-5940.
- Strzyzewska BM, Tosik A, J Crystallogr Spectrosc Res, 1991;21:379-281.
- Lin DD, Liu Y and Xu DJ. Acta Crystallogr, 2003;59:771.
- Gómez AC, Behrens B, Castro QME, et al. Castillo-Blum, Polyhedron, 2000;19:1821-1827.
- Ruiz EA, Hueso AP, Mijangos E, et al. Polyhedron, 2011;30:2090-2098
- Kim Y and Kang SK. Acta Cryst, 2015;E71:1058-1060.
- Eltayeb NE, Teoh SG, Chantrapromma S, et al. Acta Cryst, 2011; E67:m1062-m1063.
- Kim Y and Kang SK. Acta Cryst, 2015;E71:m85-m86.
- Oró J, Kimball AP. Archives of biochemistry and biophysics, 1961;94:217-222.
- Masouda MS, Soayed AA, Ali AE. Spectrochimica Acta A, 2004;60:1907-1915.
- Abd El Wahed MG and Metwally SM. Mat Chem Phys, 2002;78:299-303.
- Shaker SA, Farina Y, Mahmmod S, et al. Sains Malaysiana, 2010;39:957-966.
- Hammuda HH, Nemerb G, Sawmab W, et al. Chemico-Biological Interactions, 2008;173:84-100.
- Mikulski CM, Cocco S, Franco ND, et al. Inorganica Chimica Acta, 1985;106:89-95.
- Charland JP. Inorganica Chimica Acta, 1987;135:191-199.
- Starha P, Travnicek Z and Popa I. J Inorg Biochem, 2010;104:639-647.
- Geary W. Coord Chem Rev, 1971;7:81-85.
- Teotonio EES, Espi´nola JGP, Brito HF, et al. Polyhedron 21, 2002, 1837-1846.
- Földesi A, Trifonova A, Kundu MK, et al. Nucleotides, Nucleossides & Nucleic Acid, 2000;19: 1615-1621.
- Dhaoudi Z, Ghomi M, Mojzes P, et al. Eur Biophys, 1994;23:95-101;
- Crestoni ME and Fornarini S. J Mass Spectrom, 2003;38:854-861.
- Mostafa SI and Abd El-Maksoud S. Monatsh Chem, 1998;129:455-466.
- Savoie R, Jutier J, Prizant L, et al. Spectro chim Acta, 1982;561.
- Fujita T and T. Sakaguchi. Chem Pharm Bull. 1977;2:419-424.
- Shirotake S. Chem Pharm Bull, 1980;16:73-79.
- Masoud MS, El-Merghany A and Abd El-Kaway MY. Synthesis and Reactivity in Inorganic, Metal-Organic, and Nano-Metal Chemistry, 2009;39:537-553.
- Mostafa SI. Transition Met Chem, 2007;32:769-775.
- Mostafa SI. J Coord Chem, 2008;61:1553-1567.
- Elsayed SA, Claude BJJ, Butler IS, et al. J Molecular Structure, 2012;1028:208-214.
- El-Morsy FA, Claude BJJ, Butler IS, et al. Inorg Chim Acta, 2014;423:144-155.
- Inba PJK, Annaraj B, Thalamuthu S, et al. Bioinorganic Chemistry and Applications, 2013: 1-11.
- Elsayed SA, El-Hendawy AM, Mostafa SI, et al. Inorg Chim Acta, 2010;363:2526-2532.
- Shabana AA, Butler IS, Gilson DFR, et al. Inorg Chim Acta, 2014; 423:242-255.
- Ouf A, Ali MS, Saad EM and Mostafa SI. J Mol Struct, 2010; 973:69.
- Elsayed SA, Claude BJJ, Butler IS, et al. Transition Met Chem, 2014.
- Mostafa SI, Perlepes SP, Hadjiliadis N, et al. 2001;56:396-402.
- El-Asmy HA, Butler IS, Mouhri ZS, et al. J Mol Struct, 2014;1059:193-201.