Review
, Volume: 15( 4) DOI: 10.37532/0974-7435.2019.15(4).193Structural paradigm shift of proteins against thermal stress through evolutionary proteome
- *Correspondence:
- Anindya Sundar Panja Post Graduate Department of Biotechnology, Molecular informatics Laboratory, Oriental Institute of Science and Technology, Vidyasagar University, West Bengal, India
E-mail: biotech2ani@gmail.com
Received: July 16, 2019; Accepted: September 1, 2019; Published: September 6, 2019
Citation: Panja AS, Mandal S, Bandopadhyay B, et al. Structural Paradigm Shift of Proteins against Thermal Stress Through Evolutionary Proteome. Biotechnol Ind J. 2019;15(4):193
Abstract
The proteome body is the most important determining factors of the phenotypic characteristics of an organism. Generally a phenotypic feature physically interacts with the nature and its evolutionary changes. A sustained adverse interactions result in the extinction of the organism. The present review focuses on the different conditions like natural selection pressures, mutations, temperature etc. On the organismal genotypic features which finally dictate the proteins structure and functions. What we think as a whole, changes in natural process is basically the factors that can be termed as stress. Deviations from the existing natural cues may generate smaller or larger amount of stress on the organism. So the organism and its metabolic representative, proteins should earn the natural stress withstanding abilities for their sustenance. Structural modifications in proteins like balanced proportion of hydrophilic/ hydrophobic amino acid occurrence in its structure, presence of salt bridges and other weak interactions are responsible for structural paradism of these proteins. The peptide non-planarity is also an important determining factor that efficiently and linearly favoured the stress withstanding abilities against qualitative and quantitative stresses on the course of evolutionary period. All these factors in proteins may serve as immensely powerful force for surviving millions of years on the course of evolutionary processes
Introduction
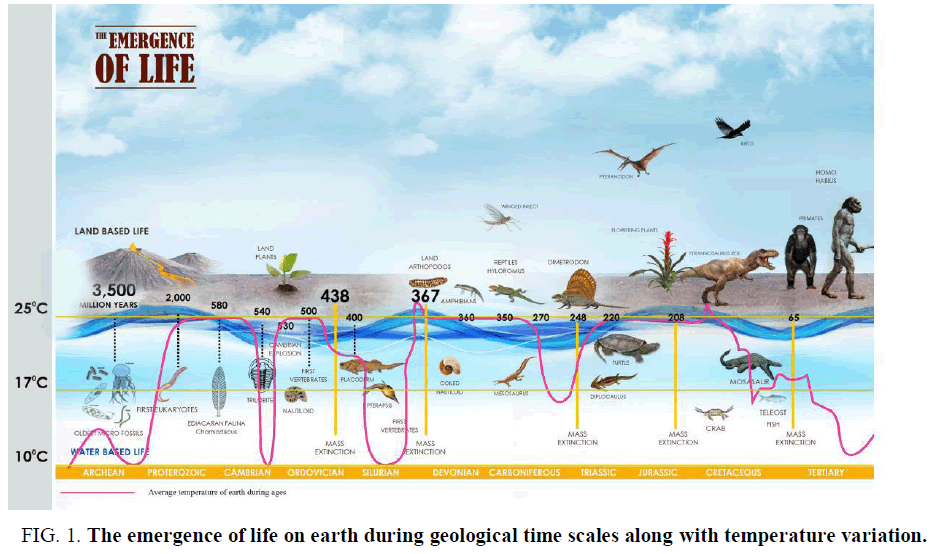
Biogenesis is a natural process where life was originated from nonliving matter such as simple organic compounds. It is thought that earth was formed about 4.5 billion years ago and the undisputed evidence of life on earth surface dates at least from3.5 billion years ago. The first evidence of life was found as a microbial mat fossil in 3.48 billion years ago from Western Australia which is thought to be single-cell prokaryotes, perhaps evolve from organic molecules surrounded by a membrane like structure called protobions [1]. The earliest microbial life on earth was dated 3.5 billion years ago, during the Eoarchean era when earth crust had solidified following the molten Hadean eon [2,3]. During the formation of first life, the temperature of earth crust had decreased and water vapour condensed into water bodies. At the transition from Archean to the Proterozoic (2.5 billons years ago), the temperature of the earth was decreased [4]. The additional heat produced during early age was due to the mixture of remnant heat from planetary accretion. The heat was generated from the core of the earth and that was produced by the decay of radioactive materials. At the beginning of Archean eon, Cyanobacterial community and Archean were the dominated flora [5]. It is believed that the Hadean started with the formation of earth at about 4.7 billion years ago and ended about 3.8 billion years ago. At the beginning of its history, earth passed the stage of melting at its external surface to the depth of several tens of kilometres. At this early age of the earth, there was no hydrosphere and the atmosphere contains no oxygen. Many volcanic activities took place on the earth's territory at that time. The earth's surface also reached a tremendous temperature up to 100000ºC and a pressure up to 109 Pa during Prebiotic history. The temperature then decrease to 10000ºC on the earth surface during the middle of the Hadean and in the Proterozoic period the temperature has declined to 400ºC, which again reduced to 220ºC in the Cretaceous period (about 100 million years ago; FIG. 1.) and the earth surface again cooled down to 150ºC at present [6].
Figure 1. The emergence of life on earth during geological time scales along with temperature variation.
Evolution of Life Through Ages
Further biological evolution of life during ages produced more complex and improved organization of life which correlated to temperature-dependent evolution in the earth [7]. Therefore, it may be concluded that the effect of temperature was unquestionably and contributed the main dynamic force of phyletic transformation or anagenesis [8].
Pleiotropic mutation events affect the evolution of gene and protein function. These duplication and divergence mechanism cause organismal fitness to depend upon the environmental stress [9]. Environmental temperature accelerates the evolutionary rate in the archaea. Temperature is the major key factor that controls the evolution dynamics but thermophilic have lower and mesophilic have higher evolutionary rate.
The Molecular Oscillator of Evolution
High variation in nucleic acids and amino acids comparison present into the mesophilic archaea is to gain the adaptation in optimal growth temperature [10]. Bacteria and Archaea show acclimatization in various environments including a wide range of temperatures [11-13]. Thermophilic water-soluble proteins including membrane proteins have increased stability at high temperature by increasing side chain burial and lower sharp turn into their structures. This structural modification has eliminated thermal sensitivity of amino acids and decrease entropy cost during the evolutionary adaptation [14]. Some particular amino acids are favoured during the protein evolution at high temperature. The corresponding gene of these favoured amino acids in thermophilic proteins contains more GC than the mesophilic one. Protein evolution is not determined by only their function and structure but also affected by various factors of gene including their position in the genome [15]. The report reveals that the thermophilic prokaryotes (Archaea and Eubacteria) adopt different strategies to maintain stability in their proteins. At DNA level, the GC-rich codons and at protein level, the charged amino acids are higher in number [16]. This favoured amino acids property made the pattern of substitutional asymmetry for free energy transfer and constitute more hydrophobicity to protect against high energy interaction [17]. Protein thermostability is an attractive field of research for its basic evolutionary as well as applied concern.
Thermostability of thermophilic proteins compared to their mesophilic homolog arises from the concomitant effects of several forces such as hydrophobic interactions, disulfide bonds, salt bridges, and hydrogen bonds. These lead to a decreased flexibility of the protein molecule by disfavouring its functionality [18]. The surface loop deletions play an important role to minimize the exposed area of protein surface [19]. Proteins from thermophilic organisms show intrinsic thermal stability but have similar structure like mesophilic homolog. Different protein families adapt to higher temperature by changing structural motifs. Due to increase of ion pairs the optimum growth temperature also increases but it has no effect on hydrogen bond and polarity. The thermophilic and extreme thermophilic proteins show stability in different growth temperatures by the presence of higher number of ion pairs, cavities, polarity and amount of all these results surface modifications. These elementary modifications into secondary structure adopted more heat withstanding ability [20]. The thermophilic proteins are able to sustain at high temperature and sufficiently show functionally stable under the extreme stress condition. Non polar glycine and isoleucine are becoming more by substitute glutamic acid and lysine on the surface of the proteins [21], which is in line with our earlier finding of lesser residue number and smaller volume of amino acids for condensing the thermophilic proteins to minimize the solvent contact, resulting evading from heat exposure [22]. Protein thermostability conferred by the protein compactness and structural rigidity is an oversimplified concept. Hence, rigidity of protein-structure may result in an unwanted decrease in its function. The overlapping zone of flexibility and rigidity in protein molecule may partially analogize to its structure-function relationship. So, it is important for the understanding and further investigation to address the discrepancies and its nullifications during protein function reservation yet attributing robust structural alteration aiming to temperature adaptation [23]. Folding and unfolding behavioural pattern proteins modified against the thermal stress by increasing Lys, Arg, and Glu and lowering the number of Ala, Asp, Asn, Gln, Thr, Ser, His [24]. The slowly evolve and more highly express proteins have been equilibrium constant for folding. The thermophilic proteins are more stable than mesophilic due to difference in amino acids composition [25]. The report reveals that ionizable residues and surface charge play important roles in protein stability, related to folding with function after burial of polar contacts and ion pairs. Charged residues further exert their profound influence by forming contact network not only with other charged residues but also with polar or nonpolar residues in the thermostable proteins [26]. Non-planer peptide bonds are responsible for adaptive evolutionary modification which may introduce an opportunity to protect protein structure against various thermal stresses. The appearance of cis-peptide bonds in these protein structures is observed more in mesophilic proteins than thermophilic ones [22]. The ionic interactions, amino acid preferences and their distribution, post-translational modifications and solute accumulation are shown to be temperature-dependent [16,22]. A large network of week interactions resulting in an extra force offers structural-rigidity, conformational entropic stability, higher ΔG in thermophilic proteins [27]. In respect to thermodynamic and kinetic stability, free energy of an enzymatic reaction is defined as ΔGstab (differentiate as folded and the unfolded states of the protein). Higher melting temperature(Tm) increases the enzyme resistance to unfolding, where 50% of the protein is in unfolded state and is usually results in irreversible inactivation/ denaturation of thermostable protein or enzyme [28-30].
Thermodynamic and kinetic stability of enzymes or protein can be defined with free energy and maybe differentiate through eight parameters, four of which are based on the change in enthalpy and entropy for protein unfolding ΔH* and ΔS* with associated convergence temperatures T*H and T*S, respectively. The rest of the other four additional parameters are associated with a rate-limiting enzyme which is scaling as constant c; enthalpy of activation ΔH++A; heat capacity change on denaturation ΔC*p; the number of amino acid residues n [31].
By increasing peptide nonplanarity of a protein, the structural stability adapted against heat-stress and more propensity of no planarity observe in C-terminal maybe validate with the fact. Nonplanarity pattern may be a determinant factor to calculate the evolutionary time scale based on the adaptations and organic evolution against stress [32].
Protein Structural Impact by Point Mutation
Point mutation is also an important factor among different perspectives for the structural and functional impact of a protein [33]. Point mutation results in different conformal changes by intermolecular bonding, globally or locally into protein structure. It may cause structural and phenotypic adaptation against thermal stress [34]. Position-specific point mutation has some clues about the evolutionary divergent line of thermophilic to mesophilic protein transition [35]. To maintain or improve the structural stability, point mutations are required which can change the amino acid sequence. In this way, over 50%-80% of all amino acids number can be changed. Although the mutations are random selection is the driving force favoured by selection pressure [36-38].
Natural mutation in protein result and change in the Gibbs free energy as; ΔG<-1 kcal/mol whereas ΔG>1 kcal/mol were considered as non-neutral which does not affect on function [39].
On the basis of the evolutionary perspective, several computational algorithms have been developed to predict the effect of a position-specific single substitute and differential amino acids propensity in proteins result structural deviation to combat against various stresses [40,41].
Evolutionary Clock Determiner
These factors may differ from taxon to taxon, both within and between species. Therefore, no single determinant has been shown to be universally attributable to protein thermostability so far [16]. In this regard, the best possible method is to study the homologous optimum thermostable proteins from different organisms, which are fighting against higher natural environmental temperature [42]. Thermophilic proteins generally have more stable folds than mesophilic proteins and it was observed that there are systematic differences in amino acid content between thermophilic and mesophilic proteins. Related correlations of amino acid frequencies with evolutionary rate and functional expression level observed within genomes [25]. Blossom is developed by Hanikoff which is based on the evolutionary rate and designed as a Dayhoff model. The substitution matrix with score has calculated by protein alignment method with all possible changes between the amino acids. In this substitution matrix 2500 protein sequences have taken for the MSA alignment among these 2000 blocks are identified from this alignment and 500 groups characterized corresponds with related proteins [43]. PAM and Blossom matrix is used to detect the homology between the protein sequences to determine the structure and function of uncharacterized proteins. This protein sequence comparison method helps to identify the network of the evolutionary root within the rapid growing protein data bank library [44]. Although Blossom and PAM matrix is not able to detail the evolutionary model whereas PMB (probability matrix from blocks) is the new model used for identifying and analyzing the evolutionary root of protein sequences [45].
Future Perspective of Thermophilic Proteome
Wilmes and bond traits define meta proteomic as “The large-scale characterization of the entire protein complement of environmental microbiota at a given point in time” [46]. Through the analysis of mass spectroscopy and proteo-bioinformatics, metaproteomic approaches successfully applied on expression of protein with metabolic function. Various protein samples are isolated from waste and ocean water as well as low diversity acid-mine drainage and biofilm on soil also [47,48]. Biomass present in the biogeochemical cycle where bacteria and archaea play the key role, it is difficult to identify the marine microorganism how they operate bio-geochemical processes during their marine life. Meta genomics and meta-proteomics studies help to find out the metabolic potential of microorganism of the environment [49-53]. Amino acids composition may help to predict the thermostability or metastability of a protein by using PSD tool. Thermozymes are used in various biotechnological and industrial applications such as Petroleum, Chemical, Pulp and Paper industries to eliminate environmental hazards [54]. Thermo enzymes are uniquely stable against high temperature and pH, so by replacing mesophilic enzyme they are used in industrial process conditions [55]. The enzyme used in the industrial process mainly isolated from mesophiles because the thermophilic enzymes are limited. Thermostable enzymes are mainly isolated from thermophilic organisms. These have been identified and isolated from different exotic ecological zone of the earth [56,57].
Discussion and Conclusion
The temperature variation is one of the major factors that lead to evolutionary rates into the organisms. The amino acid substitutional information provides an opportunity to investigate the functional variation of protein during evolutionary history. High substitution rates of amino acids are observed in all the lineages both thermophilic, mesophilic as well as psychrophilic proteins. The plasticity of protein structural diversity may help to interpret accurately the results of phyloinformatics data of both thermophilic and mesophilic proteins.
Acknowledgment
This work was supported by the Oriental Institute of Science and Technology in the frame of our study.
Compliance with Ethical Standards
This article is a review, so it does not contain studies with human or animal subjects performed by any of the authors that should be approved by Ethics Committee.
Competing Interests
Anindya Sundar Panja, Shiboprosad Mandal, Bidyut Bandyopadhyay, and Smarajit Maity confirm that this article content has no conflicts of interest.
Contributions
A.S.P. wrote the manuscript with contributions from S.P, B.B. and S. M. All authors approved the manuscript before submission.
References
- Zimmer C. Origins. On the origin of eukaryotes. Science. 2009;325:666-8.
- Wilde SA, Valley JW, Peck WH. Evidence from detrital zircons for the existence of continental crust and oceans on the Earth 4.4 Gyr ago. Nature. 2001;409:175-8.
- Schopf JW, Kudryavtsev AB, Agresti DG, et al. Laser-Raman imagery of Earth's earliest fossils. Nature. 2002;416:73-6.
- Dean Tim. World’s oldest fossils reveal earliest life on Earth. Aust Life Sci. 2011.
- Nisbet EG, Fowler. Archaean metabolic evolution of microbial mats. Proceedings of the Royal Society. Biology. 1999;266:2375.
- Novikov VP. The three-stage origin of life as a result of directional darwinian evolution, Cornell University library. Quantitative Biology. 2012.
- Henderson LJ. The fitness of the environment (Macmillian, New York), 1913.
- Waring GA. Survey Prof Paper. 1965:492.
- Misha S, Dan ST. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2011;11:572-82.
- Mathieu G, Manolo G. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in Archaea. Mol Biol Evol. 2011;28:2661-74.
- Boussau B, Gouy M. Efficient likelihood computations with nonreversible models of evolution. Syst Biol. 2006;55:756-68.
- Galtier N, Tourasse N, Gouy M. A non hyperthermophilic common ancestor to extant life forms. Science. 1999;283:220-21.
- Gaucher EA, Govindarajan S, Ganesh Palaeotemperature trend for Precambrian life inferred from resurrectedproteins. Nature.2008;451:704-7.
- Meruelo AD, Han SK, Kim S, et al. Structural differences between thermophilic and mesophilic membrane proteins. Protein Sci. 2012;21:1746-53.
- Csaba P, Balazs P, Martin JL. An integrated view of protein evolution. Nat Rev Genet. 2006;7:337-48.
- Trivedi S, Gehlot HS, Rao SR. Protein thermostability in Archaea and Eubacteria. Genet Mol Res. 2006;5:816-27.
- John HM. Temperature adaptation at homologous sites in proteins from nine thermophile-mesophile species pairs. Genome Biol Evol. 2010;2:267-76.
- Yu H, Huang H. Engineering proteins for thermostability through rigidifying flexible sites. Biotechnol Adv. 2014;32:308-15.
- Kumar S, Nussinov R. How do thermophilic proteins deal with heat? Cell Mol Life Sci. 2001;58:1216-33.
- Andras S, Peter Z. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits. Structure. 2000;8:493-504.
- Mizuguchi K, Sele M, MV Cubellis. Environment specific substitution tables for thermophilic proteins.BMCBioinformatics. 2007;8:15.
- Panja AS, Bandopadhyay B, Maiti S. Protein Thermostability is owing to their references to non-polar smaller volume amino acids, variations in residual physico chemical properties and more salt-bridges. PLoS One. 2015;10:e0131495.
- Karshikoff A, Nilsson L, Ladenstein R. Rigidity versus flexibility: The dilemma of understanding protein thermal stability. FEBS J. 2015;282:3899-917.
- Anna VG, Sergiy Garbuzynskiy, Michail YL, et al. Different packing of external residues can explain differences in the thermostability of proteins from thermophilic and mesophilic. Bioinformatics. 2007;23:2231-8.
- Joshua L, Cherry. Highly expressed and slowly evolving proteins share compositional properties with thermophilic proteins. Mol Biol Evol. 2010;27:735-41.
- Chen C, Li L, XiaoY. All-atom contact potential approach to protein thermostability analysis. Biopolymers. 2007;85:28-37.
- Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Sci. 2006;5:1569-78.
- Bommarius AS, Paye MF. Stabilizing biocatalysts. Chem Soc Rev. 2013;42:6534-65.
- Vieille C, Zeikus GJ. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1-43.
- Sanchez-Ruiz JM. Protein kinetic stability. Biophys Chem. 2010;148:1-15.
- Corkrey R, McMeekin TA, Bowman JP, et al. Protein Thermodynamics can be predicted directly from biological growth rates. PLoS ONE. 2014;9(5):e96100.
- Smarajit Maiti, Anindya Sundar Panja, Bidyut Bandopadhyay. Higher peptide nonplanarity (u) close to protein carboxy-terminal and its positive correlation with dihedral-angle is evolved conferring protein thermostability, Progress in Biophysics and Molecular Biology. 2018.
- Schaefer C, Rost B. Predict impact of single amino acid change upon protein structure. BMC Genomics. 2012;13.
- Chasman D, Adams RM. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J Mol Biol. 2001;307:683-706
- Sunyaev S, Lathe W, Bork P. Integration of genome data and protein structures: Prediction of protein folds, protein interactions and ‘‘molecular phenotypes’’ of single nucleotide polymorphisms. Curr Opin Struct Biol. 2001;11:125-30.
- Shakhnovich EI, Gutin AM. Influence of point mutations on protein structure: the probability of a neutral mutation. Journal of Theoretical Biology. 1991;149(4):537-546.
- Sander C, Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9(1):56-68.
- Rost B. Twilight zone of protein sequence alignments. Protein Engineering. 1999;12(2):85-94.
- Christian Schaefer, Burkhard Rost. Predict the impact of single amino acid change upon protein structure. BMC Genomics. 2012;13;S4.
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863-74.
- Anindya Sundar Panja, Bidyut Bandopadhyay, Akash Nag, et al. Protein Secondary Structure Determination (PSSD): A new and simple approach. Current Proteomics. 2019;16:246.
- Pucci F, Rooman M. Towards an accurate prediction of the thermal stability of homologous proteins. J Biomol Struct Dyn. 2015;33:1-11.
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. PNAS. 1992;22:10915-19.
- Gavin A, Price E, Crooks RE, et al. Statistical evaluation of pairwise protein sequence comparison with the Bayesian bootstrap. Bioinformatics. 2002;21:3824-31.
- Shalini V, Andrew S, Elisabeth RM. A Transition probability model for amino acid substitutions from blocks. J Bioinform Comput Biol. 2003;6:997-1010.
- Wilmes P, Bond PL. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ Microbiol. 2004;6:911-20.
- VerBerkmoes NC, Denef VJ, Hettich RL, et al. Functional analysis of natural microbial consortia using community proteomics. Nat Rev Microbiol. 2009;7:96-205.
- Schneider T, Riedel K. Environmental proteomics: Analysis of structure and function of microbial communities. Proteomics. 2010;10:785-98.
- Hettich RL, Sharma R, Chourey K, et al. Microbial metaproteomics: identifying the repertoire of proteins that microorganisms use to compete and cooperate in complex environmental communities. Curr Opin Microbiol. 2012;15:373-80.
- DeLong EF, Preston CM, Mincer T, et al. Community genomics among stratified microbial assemblages in the ocean′s interior. Science. 2006;311:496-503.
- Bruins ME, Janssen AE, Boom RM. Thermozymes and their applications: A review of recent literature and patents. Appl Biochem Biotechnol. 2001;90:155-86.
- Tyson GW, Chapman J, Hugenholtz P, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37-43.
- Yooseph S, Sutton G, Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16.
- Panja AS, Akash N, Bandopadhyay B, et al. Protein Stability Determination (PSD): A tool for proteomics analysis. Current Bioinformatics. 2018;13:1-9.
- Bahrami A, Shojaosadati S, Mahbeli G. Biodegradation of dibenzothiophene by thermophilic bacteria. Biotechnol. Lett. 2001.23:899-901.
- Bauer M, Driskil L, Callen W, et al. An endoglucanase EglA, from the hyperthermophilic archaeon Pyrococcus furiosus hydrolyzes a-1,4 bonds in mixed linkage (1-3), (1-4)-b-D-glucans and cellulose. J Bacteriol. 1999;18:284-90.
- Antranikian G, Herzberg C, Gottschalk G. Production of thermostable a-amylase, pullulanase and a-glucosidase in continuous culture by a new Clostridium isolate. Appl Environ Microbiol. 1987;53:1668-73.