Original Article
, Volume: 15( 1)Strain Improvement of Halotolerant Actinomycete for Protease Production by Sequential Mutagenesis
- *Correspondence:
- Payal Mehtani Department of Biotechnology, The IIS University, Jaipur, India, Tel: 9783307166; E-mail: payal.mehtani@iisuniv.ac.in
Received: March 2, 2017 Accepted: March 17, 2017 Published: March 22, 2017
Citation:Mehtani P, Sharma C, Bhatnagar P. Strain Improvement of Halotolerant Actinomycete for Protease Production by Sequential
Mutagenesis. Int J Chem Sci. 2017;15(1):109.
Abstract
Proteases are the natural biocatalysts that due to their enormous range and specificity have attracted the attention of the scientists globally for better microbial sources for enhanced production of the enzyme. However, bacterial proteases from Bacillus are comprehensively studied and characterized, less attention has been paid to proteases produced by Actinomycetes. In the present investigation, a protease producing halotolerant and alkaliphilic Actinomycete, A1F2, was isolated from water of Sambhar Salt Lake, Rajasthan, India identified as Streptomyces sp. To further enhance the production, the isolate was subjected to sequential random mutagenesis using acridine orange, ethidium bromide and Ultraviolet radiation. The protease production in shake flask increased approximately 2.2-fold from parent to the final mutant. The results suggest that the process of sequential random mutagenesis is more efficient in strain improvement than a single mutagen treatment. The hyper active protease producing mutant can be used for industrial applications.
Keywords
Acridine orange; Ethidium bromide; Protease; Sequential random mutagenesis; Ultraviolet radiation
Introduction
Proteases are enzymes that conducts proteolysis, alone accounts for 65% [1] of the total worldwide sale of enzymes among other two largest groups of industrial enzymes viz., lipases, amylases, and has increased from 59% reported by Rao et al. [2]. Proteases are exploited in various industries like detergent, leather, silk manufacturing, food, pharmaceutical industry and even in bioremediation processes [3]. Among all types of proteases, alkaline proteases find maximum use in industries [2]. Many researchers have also reported the applications of alkaline proteases in hydrolysis of fibrous proteins of horn, feather and hair [4,5], in recovery of silver from X-ray films by hydrolysis of gelatin [6-8].
While proteases from bacteria are extensively characterized, less attention has been paid to proteases produced by Actinomycetes. Actinomycetes have been majorly studied extensively for the production of antibiotics [9,10]. The use of Actinomycetes for enzyme production is now being explored on a large scale [11].
Mesophilic and extremophilic Actinomycetes isolated from soil of different areas of Georgia showed protease activity [12]. In India, members of the genus Micromonospora and Streptomyces, isolated from Pudimadaka coast of the Bay of Bengal showed proteolytic activities and thus have provided an immense potential of finding industrially important enzymes from such halophilic habitats [13]. An alkalophilic Actinomycetes showing proteolytic activity was also reported from Saudi Arabia [14].
There is a growing demand in industries for hyper protease producing microbial strains. The yield of protease production depends on the type of carbon and nitrogen sources in the medium. Therefore, the production can be enhanced by optimizing these nutrient sources along with other culture conditions. Also, mutants can be generated using physical and chemical mutagens to improve the efficiency of microbes for protease production [15] which would also reduce cost of production. Microbial strain improvement by conventional mutagenesis (UV or chemical exposure) is well known. In the present study, sequential random mutagenesis was used to enhance protease production from an Actinomycete isolate.
Materials and Methods
Isolation of Actinomycete strain
A halotolerant and alkaliphilic Actinomycete, A1F2, was isolated from waters of Sambhar Salt Lake, Rajasthan, India. The isolation was carried in Actinomycete Isolation Agar (AIA) media containing cyclohexamide (50 μg/ml), nystatin (25 μg/ml) and NaCl (3%) at 28°C.
Screening for proteolytic activity
The isolate was streaked on milk agar media containing (g/l): 33.33 skim milk powder, 1.18 potassium dihydrogen phosphate, 1.0 disodium hydrogen phosphate, 1.25 glycine, 0.66 sodium hydroxide, 30.0 sodium chloride, 16.67 agar, adjusted to pH 8 and incubated for 48 h at 28°C and observed for zone of hydrolysis.
Phenotypic characterization
The organism was identified and characterized by following the standard protocols mentioned in Bergey’s Manual of Determinative Bacteriology [16].
Protease production in liquid-batch fermentation
The culture was grown in 250 ml of Erlenmeyer flasks containing 100 ml medium consisting of (g/1000 ml) dextrose 30.0, sodium nitrate 3.0, yeast extract 4.0, magnesium sulphate 0.5, di-potassium hydrogen phosphate 1.0 and calcium carbonate 1.0 for 48 h at 28°C at 160 rpm on a shaking incubator (MAC). Supernatant was collected after centrifugation at 10,000 rpm for 10 minutes at 4°C in a cooling centrifuge (Sigma Sartorius 1-15 PK).
Protease assay
The proteolytic activity of the crude supernatant was determined using spectrophotometric method [17]. One unit of enzyme activity was defined as the amount of enzyme required to liberate one micromole tyrosine equivalents from casein per minute under the experimental conditions used.
Molecular identification by 16srDNA sequencing
DNA was isolated from 24 h old culture and fragment of 16S rDNA gene was amplified by PCR using 8F and 1492R primers. Forward and reverse DNA sequencing reaction of PCR amplicon was carried out using BDT v3.1 Cycle sequencing kit on ABI 3730xl Genetic Analyzer. The consensus sequence of 1414 bp 16S rDNA gene was generated from forward and reverse sequence data using aligner software that was used to carry out BLAST with the NR database of NCBI genbank database. Based on maximum identity score first ten sequences were selected and aligned using multiple alignment software program Clustal W. Distance matrix was generated using RDP database and the phylogenetic tree was constructed using MEGA 4.
Strain improvement by random mutagenesis
Acridine orange (AO) treatment: Two-day old culture of the parent strain was inoculated in 50 ml of M1 medium and incubated at 28°C for 48 h at 160 rpm. 2 ml of culture was added to 1 ml of AO dilutions (10-1 μg/ml to 105 μg/ml). After 90 minutes’ incubation, 2 μl of it was plated on milk agar plates. Untreated parent strain was taken as control. The plates were incubated at 28°C for 48 h and observed for colony formation and hydrolysis. Plates showing 99% killing were used for mutant selection based on morphological changes and zone of hydrolysis.
Ethidium bromide (EtBr) treatment: Mutants selected from AO treatment were further subjected to mutagenesis by EtBr (0.2 g/l to 1.5 g/l) using the above method. AO treated mutant was taken as control.
UV irradiation: Mutants selected after EtBr mutagenesis were inoculated in M1 medium for 24 h under same conditions. The cultures were centrifuged at 4°C and 5000 × g for 10 minutes. Supernatant was slowly decanted and the pellet was washed twice by cold sterile 0.9% NaCl and resuspended in 50 ml of the same [18]. 10 ml aliquots of cell suspension were transferred to sterile petri dishes and irradiated with UV radiations at 254 nm from a distance of 10 cms in a UV chamber (ACCO) for a period of 1-4 minutes. Photoreactivation was avoided by keeping the plates overnight in dark by wrapping in aluminium foil. EtBr treated mutants was taken as control. Irradiated cell suspension was centrifuged at 5000×g for 10 minutes and pellet was re suspended in 10 ml of M1 medium and incubated at 28°C for 18 h. Cultures were serially diluted in sterile 0.9% NaCl solution and spread on milk agar plates, incubated at 28°C for 48 h. Mutant selection procedure was similar to AO and EtBr treatment.
After obtaining the mutants from the above experimentation, each mutant was streaked on milk agar plates and compared with the parent strain for diameter of zone of hydrolysis after incubation of 48 h.
Statistical analysis
Protease activity is presented as mean ± standard deviation (SD). One-way Analysis of variance of (ANOVA) was used to test differences among proteolytic activity of parent and mutant strains.
Results
Isolation, screening and characterization of proteolytic Actinomycete
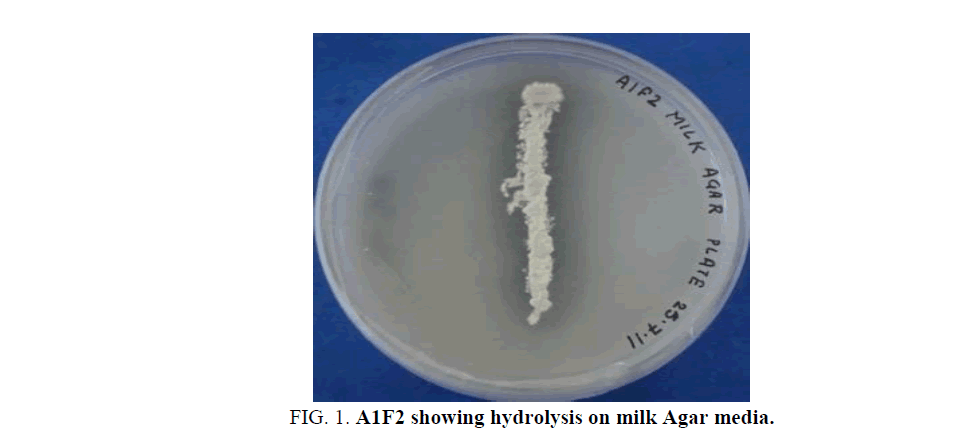
Isolated Actinomycete strain, A1F2, showed a clear zone of hydrolysis on milk Agar media (Figure. 1). The isolate was found to be Gram positive, non-acid-fast, filamentous bacteria having a white coloured aerial mycelium. The phenotypic characterization of the isolate showed that it may belong to the genus Streptomyces.
Figure 1: A1F2 showing hydrolysis on milk Agar media.
Molecular identification by 16srDNA sequencing
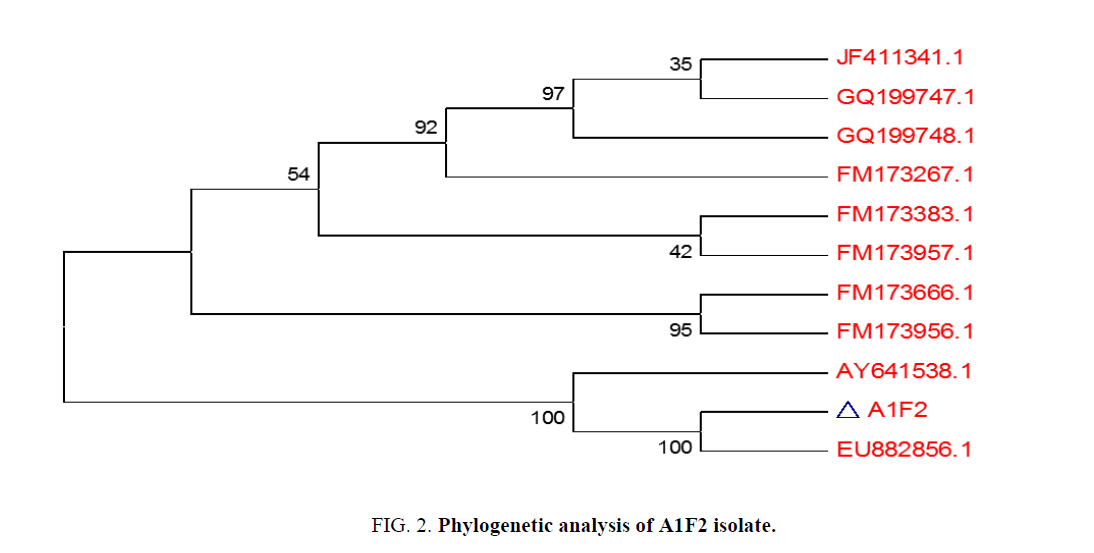
99% homology of the isolate A1F2 was found to be with Streptomyces sp. LD48 (Gen Bank Accession Number: AY641538.1) as derived from nucleotide homology and phylogenetic analysis (Figure. 2).
Effect of AO, EtBr and UV mutagenesis
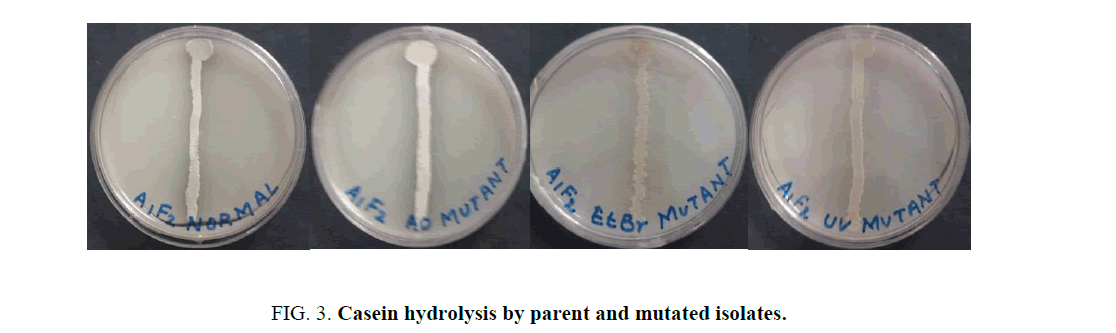
When the isolate was subjected to mutagenesis with AO, EtBr and UV, an increment in the diameter of zone of hydrolysis was observed at each step on milk agar plates (Figure. 3).
Protease assay
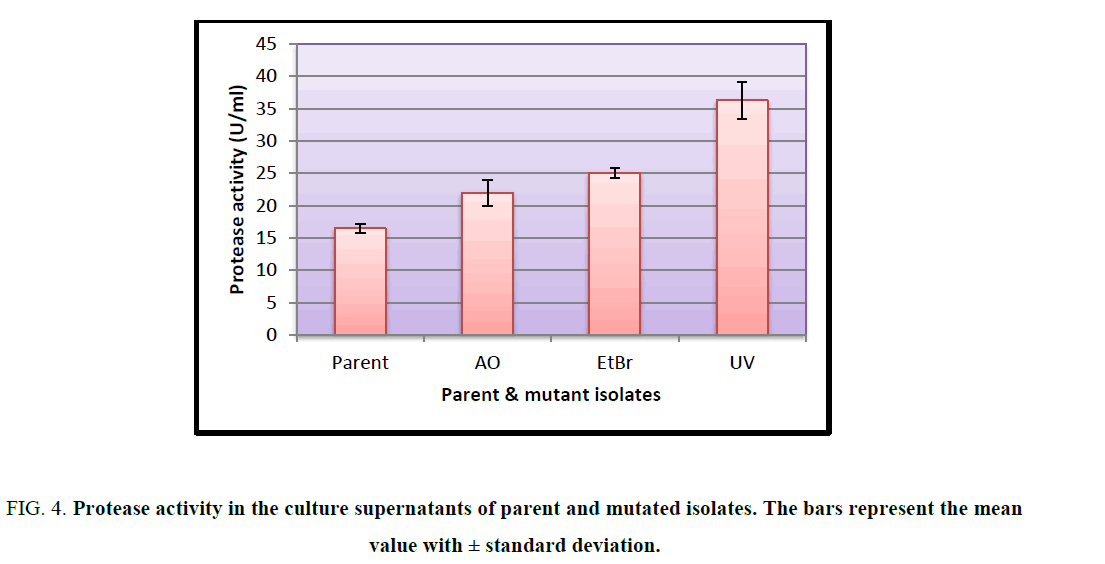
Sequential random mutagenesis increased protease activity at each step (Figure. 4) Protease activity of parent strain was least 16.448 U/ml ± 0.6625 U/ml, which increased on mutagenesis with acridine orange (21.912 U/ml ± 1.983 U/ml), then further with ethidium bromide (25.049 U/ml ± 0.721 U/ml) and highest activity was obtained with UV mutant as 36.230 U/ml ± 2.804 U/ml. There was a significant difference in the activity of the parent and mutant isolates at 1% level of significance as determined by ANOVA (F=65.44).
Figure 4: Protease activity in the culture supernatants of parent and mutated isolates. The bars represent the mean value with ± standard deviation.
Discussion
Mutations play an important role in improving the microbial strain for protease production and thereby increase the yield of alkaline protease. Several mutational techniques are being used for strain improvement to produce maximum yield of enzyme [19]. The current study was aimed at characterizing a protease producing Actinomycete from Sambhar Salt Lake and to increase the production of protease by employing random sequential mutagenesis. The isolate was identified as a Streptomycetes sp. Sequential mutagenesis was conducted by selecting a relatively stable and higher-yielding protease mutant strain, isolated from the previous treatment, for the next mutagenesis step. An increment in the diameter of zone of hydrolysis on milk agar plates at each step was observed when the isolate was subjected to mutagenesis with AO, EtBr and UV. These mutants were further evaluated through shake flask enzyme production studies over the parent isolate. It was observed that after each mutagenic step protease activity also increased (Figure. 4). Among all the mutants so obtained, the maximum protease production (36.230 U/ml ± 2.804 U/mL) was observed for the UV mutant. The results showed that the UV mutant was hyper-active mutant, giving approximately 2.2-fold more protease activity over the parent strain (16.448 U/ml ± 0.6625 U/mL) in similar culture conditions. The findings of present study are consistent with the previous studies carried out by Shikha and Darmwal [20] who have reported 1.44-fold increase in protease production over the parent strain of Bacillus pantotheneticus while Azad [21] reported thousands fold increase in enzyme activity after mutagenic treatment in Bacillus isolate MA6. Dutta and Banerjee [22] also observed 2.5-fold increase in alkaline protease production by UV mutant Pseudomonas sp. JNGR242. It can be concluded from the present study that sequential random mutagenesis improved protease production from Streptomycetes sp. Also, the results indicate that the sequential mutagenesis process was more effective in improving the yield of alkaline protease as compared to a single mutagen treatment. The mutant hence obtained can be exploited commercially for large scale industrial production of proteases.
Acknowledgement
Authors gratefully acknowledge Department of Science and Technology, Rajasthan, India for providing the financial assistance for this work (Grant no. P7 (3) DST/R&D/2008/10775) and The IIS University, Jaipur for the facilities and financial support.
References
- Aqel H. Phenotypic and protease purification of two different thermophilic Bacillus strains HUTBS71 and HUTBS62. Ann Biol Res. 2012;3:1747-56.
- Rao MB, Tanksale AM, Ghatge MS, et al. Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev. 1998;62:597-635.
- Mehtani P, Sharma C, Chatterjee S, et al. Proteases: The industrial biocatalyst, IISU-JOST. 2013;2:1-12.
- Anwar M. Saleemuddin, Alkaline Protease: A review. Bioresour Technol. 198;6:175-83.
- Giongo JL, Lucas FS, Casarin F, et al. Keratinolytic protease of Bacillus species isolated from the Amazon basin showing remarkable dehairing activity. World J Microb Biot. 2007;23:375-82.
- George S, Raju V, Krishnan MR, et al. Production of protease by Bacillus amyloliquefaciens in solid state fermentation and its application in the unhairing of hides and skins. Process Biochem. 1995;30:457-62.
- Singh J, Vohra RM, Sahoo DK, et al. Alkaline protease from a new obligate alkalophilic isolate of Bacillus sphaericus. Biotechnol Lett. 1999;21:921-24.
- Pathak AP, Deshmukh KB. Alkaline protease production, extraction and characterization from alkaliphilic Bacillus licheniformis KBDL4: A Lonar Soda Lake isolate. Indian J Exp Biol. 2012;50:569-76.
- Hamm DA, Fiedler HP, Goodfellow M, et al. An antibiotic, antiparasitic and antitumor compound produced by a novel alkaliphilic Streptomyces strain. J Antibiot. 2003;56:639-46.
- Ellaiah P, Raju KV, Adinarayana K, et al. Bioactive rare Actinomycetes from indigenous natural substrates of Andhra Pradesh. Hindustan Antibiot Bull. 2002;44:17-24.
- Syed DG, Agasar D, Pandey A. Production and partial purification of α-amylase from a novel isolate Streptomyces gulbargensis. J Ind Microbiol Biotechnol. 2009;36:189-94.
- Gurielidze M, Pataraya D, Cholokava N, et al. Extremophilic Actinomycetes, distributed in various types of soils of Georgia and their protease activity. Bull Georg Natl Acad Sci. 2010;4:81-5.
- Haritha R, Siva Kumar K, Jagan Mohan YSYV, et al. Amylolytic and proteolytic Actinobacteria isolated from marine sediments of Bay of Bengal. Int J Microbiol Res. 2010;1:37-44.
- Ara NA, Bukhari DR, Wijayanti MA, et al. Proteolytic activity of alkaliphilic, salt-tolerant Actinomycetes from various regions in Saudi Arabia. Afr J Biotechnol. 2012;11:3849-57.
- Wang HY, Liu DM, Liu Y, et al. Screening and mutagenesis of a novel Bacillus pumilus strain producing alkaline protease for dehairing. Lett Appl Microbiol. 2007;44:1-6.
- Holt JG, Krieg NR, Sneath PHA, et al. (eds.), Bergey's Manual of Determinative Bacteriology, Lippincott Williams & Wilkins, A Wolters Kluwer Company. 2000;9-12.
- Cupp-Enyard C. Sigma's non-specific protease activity assay-casein as a substrate. J Vis Exp. 2008;19:1-2.
- Kamal F, Samadi N, Assadi MM, et al. Mutagenesis of Leuconostoc mesenteroides and selection of dextransucrase hyperproducing strains, DARU. 2001;9:18-23.
- Chand P, Aruna A, Maqsood AM, et al. Isolation of cellulase producing fungi and increasing cellulose production using novel mutations. J Appl Micrbiol. 2005;98:318-23.
- Shikha SA, Darmwal NS. Improved production of alkaline protease from a mutant of alkalophilic Bacillus pantotheneticus using molasses as a substrate. Bioresour Technol. 2007;98:881-85.
- Azad AK. Factors influencing the production of alkaline protease by a Bacillus isolate MA6 and its applications in hide processing. 1994.
- Dutta JR, Banerjee R. Isolation and characterization of a newly isolated Pseudomonas mutant for protease production. Brazil Archiv Biol Biotech. 2006;49:37-47.