Original Article
, Volume: 14( 4)Production, Optimization and Partial purification of Acid Protease from Aspergillus spp. Isolated from Soil Contaminated with Abattoir Waste
- *Correspondence:
- G. Narasimha, Applied Microbiology Laboratory, Department of Virology, Sri Venkateswara University, Tirupati, AP, India, E-Mail: dr.g.narasimha@gmail.com
Received: June 24, 2016; Accepted: June 29, 2016; Published: July 07, 2016
Citation: Ashraf G. Real Time Based RT-PCR Detection of DUF538 Gene Expression in Drought-Challenged Celosia. Biochem Mol Biol 2016; 2(1): 101.
Abstract
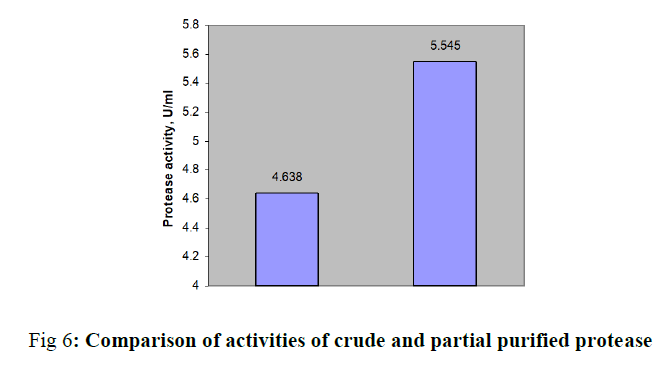
Acid protease has applications in pharmaceutical, food, leather, detergent industries. In this study, we performed medium optimization for protease production through one variable at a Time. Submerged fermentation of fungal culture Aspergillus was initiated with equal volumes of molasses and cheeses whey. The optimal pH and time of incubation was carried over pH range 2 to 9 and time of 3 to 7 days. Further enzyme productivity was tested with 1% (w/v) nitrogen sources followed by carbon sources. Best carbon and nitrogen sources were optimized upto 3% (w/v). The crude protease was partially purified by performing salt precipitation over 40 to 85% saturation. The protease activity of 3.648 μ/ml was obtained with the optimal pH of 4.5 and an incubation of 5 days. Best additional nitrogen source was found to be soya bean meal and its optimum concentration was determined as 1% (w/v). Similarly glucose enhanced the enzyme activity from 4.171 to 4.638 U/ml and finally the optimal values were found to be 1.5% (w/v). Production medium of acid protease from Aspergillus spp. was designed through conventional method, One Variable At a Time (OVAT) in an easy manner. Peak enzyme activity was found to be 4.638 U/ml with the combination of molasses and cheese whey supplemented with 1% (w/v) soya bean meal and 1.5% (w/v) of glucose. The protease was partially purified through precipitation by ammonium sulphate followed by dialysis with an enhanced the activity of 5.545 μ/ml.
Keywords
Acid protease, Aspergillus spp., Optimization, Partial purification
Introduction
Proteases are important industrial enzymes due to their enormous applications in pharmaceutical, food, leather, detergent, and other biotechnology industries and these accounts for 60% of total enzyme sales [1,2]. These biocatalysts are classified as acid, neutral, and alkaline proteases based upon the pH at which they present maximum activity[3]. Acid protease from fungal species have been exploiting for more industrial applications [4].
Submerged fermentation (SmF) is preferably employed for the production of proteases [5]. In this process microbial cells and media components are present in the submerged state in the liquid medium with a large quantity of aqueous solution [6]. Since the contents are in submerged state the transfer of heat and mass is more efficient, and is amenable for modeling the process. The scaling up of the process is very easy. Downstream processing of enzymes is achieved through various unit operations such as centrifugation, filtration, precipitation, solvent extraction, membrane separation processes, chromatography, and supercritical fluid extraction [7].
Molasses, a by-product of sugar industry, is an attractiv7e fermentation medium since main constituents are nutrients and minerals that promote microbial growth. Moreover, it is a low cost raw substance as well as abundantly available [5]. Similarly cheese whey, effluent of dairy industry is a good source of protein and can replace commercial available nitrogen sources. Taking into consideration, the increasing demand for proteases and the need for economical proteases of commercially useful acid proteases from novel sources, present study was carried to optimize the cultural conditions for the production of acid protease by the SmF by Aspergillus spp. isolated from soil contaminated with abattoir wastes using OVAT and to purify the acid protease by ammonium sulphate precipitation methods.
Materials and Methods
Materials
All the chemicals used in this investigation were of analytical grade. Molasses and cheese whey were procured from local sugar and dairy industries.
Fungal culture
The fungal culture, Aspergillus spp. used in this study was isolated from soil contaminated with abattoir waste contaminated soil [5].
Submerged fermentation (SmF)
Local procured molasses was highly viscous brown colored solution which was diluted ten times to made it amicable to production. It was sterilized at 121°C, 15 psi for 20 minutes. Protein source, Cheese whey, was sterilized by exposing it to UV radiation for 30 minutes in order to avoid denaturation of protein content of whey during sterilization. Fermentation was carried out in 250 ml Erlenmeyer flask with 1: 1 ratio of sterilized molasses and cheese whey and then inoculated with 5% (v/v) conidial suspension of Aspergillus spp. Further flask was transferred to shaker and incubated for seven days 150 rpm, a 320C. Later, the contents of the flask were filtered through Whatman No.1 filter paper and the filtrate was used for the assay of protease [8]. Activity of protease was expressed as μ/ml of enzyme. One unit of protease is defined as the amount of enzyme that releases one μmole of tyrosine / min under assay conditions. Protein was analyzed by Lowry method [9].
Effect of pH
Influence of initial pH of fermentation medium on growth of Aspergillus spp. and enzyme productivity was studied with pH range of 2 to 9.
Time of incubation
The time of incubation was determined by carrying out the fermentation for 3, 4, 5, 6 and 7 days with medium consisting of equal volumes of molasses and cheese whey at temperature of 32°C.
Influence of nitrogen sources on protease production
The effect of nitrogen sources on acid protease production was studied using the medium containing 25 ml of molasses, 25 ml of cheese whey and was supplemented with 1% (w/v) of different nitrogen sources such as potassium nitrate, sodium nitrate, ammonium nitrate, casein, beef extract, peptone, yeast extract and soya bean meal. The experiment was further carried out with different concentrations of soy bean meal ranging from 0. 5-6% (w/v).
Role of carbon sources on the activity of enzyme.
The effect of carbon sources on protease production was studied using media containing (1%) glucose, mannitol, sucrose, lactose, cellulose and starch. The study was carried out with different concentrations of glucose to determine the optimum concentration of glucose in the range of 0.5 to 3% in SmF.
Partial purification of acid protease
For the purification of crude enzyme from fermentation broth, first filtrate was centrifuged at 4000 rpm for 45min at 40C then protein mixture was recovered from the supernatant by salt precipitation. For this, ammonium sulphate was added slowly to the 25 ml of crude enzyme source with constant stirring. The concentration of salt was increased in 5% increment from 40 to 85% saturation in order to precipitate the protein. The resulted precipitate was separated by centrifugation at 3000 rpm for 30 min at 40°C. The solid pellet was dissolved in 50 mM Tris-HCl (pH 5.5). Resulted protein mixture was desalted through dialysis for 24 hrs using nitrocellulose membrane with MWCO < 10 KDA. Sample was analyzed for protease in each step of purification.
Result and Discussion
Extracellular acid protease was produced in SmF of local soil isolate Aspergillus spp. with molasses as main carbon supplement while cheese whey was served as main nitrogen source.
Effect of pH on enzyme activity
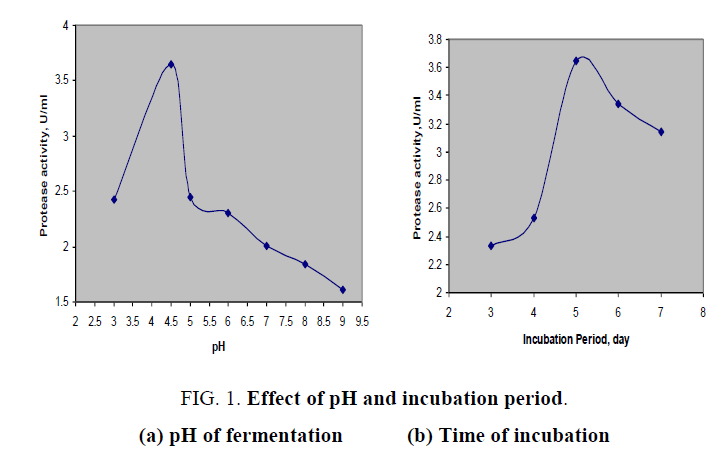
The effect of pH on protease production was shown in FIG. 1a. From this plot, it was noticed that initial of fermentation medium had played a vital role in the production of present biocatalyst and activity of enzyme was raised to maximum at pH of 4.5 and then decreased continuously with the increase in initial pH of fermentation medium. Hence the optimal pH was found to be at 4.5 as the Aspergillus spp. secreted the maximum enzyme with an activity of 3.648 U/ml. It was confirmed that isolated fungal culture was able to produce acidic protease. Protease activity of 1.516 U/ml was obtained over pH higher than neutral value. According to previous study [10] physico-chemical properties of aspartic protease, a kind of acidic protease from Onopordum ecanthium, and reported that it has exhibited its maximum activity at pH 4.
FIG. 1. Effect of pH and incubation period.
(a) pH of fermentation (b) Time of incubation
Impact of cultivation time
The time course for the production of protease was studied in shake flasks (FIG. 1b) over operating range of 3 to 7 days of incubation of medium at 32°C. The enzyme production was gradually increased with progress in cultivation time and highest protease activity was observed at fifth day of incubation. After that, the enzyme production was decreased could be due to unavailability of nutrients to fungal cultures. Similar studies were performed for acid protease from Aspergillus foetidus in a controlled fermentation [11].
Effect of nitrogen sources
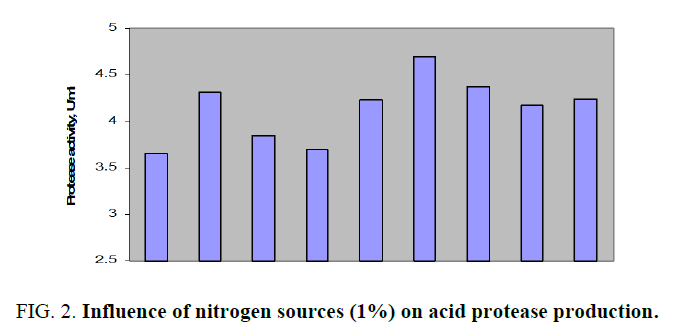
Results of impact of various nitrogen sources on acid protease production by Aspergillus spp. were depicted in FIG. 2.
FIG. 2. Influence of nitrogen sources (1%) on acid protease production.
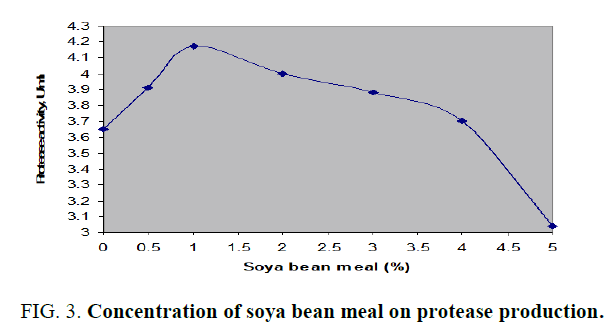
All the above mentioned additional nitrogen sources to main nitrogen source, cheese whey, had successfully increased the acid protease amount. The production was better with organic nitrogen sources than with inorganic sources. The protease activity for above mentioned nitrogen sources was in the range of about 3.648 to 4.316 U/ml. Peak enzyme production was observed with organic casein while the lowest activity with ammonium nitrate. Soya bean meal was considered as the effective additional nitrogen source in order to economize the fermentation medium as its protease activity was 8% less than that of casein. Further, the effect of various concentrations of soya bean meal on acid protease production by Aspergillus spp. in SmF was shown in FIG. 3. From the above results it was observed that 1% (w/v) soya bean meal had shown maximum protease activity of 4.171 U/ml. Similar studies were carried out alkaline protease production from Bacillus subtilis with the design of production medium with dual nitrogen sources, casein and peptone and noticed augmented enzyme activity with additional nitrogen source, peptone [12].
FIG. 3. Concentration of soya bean meal on protease production.
Additional carbon sources
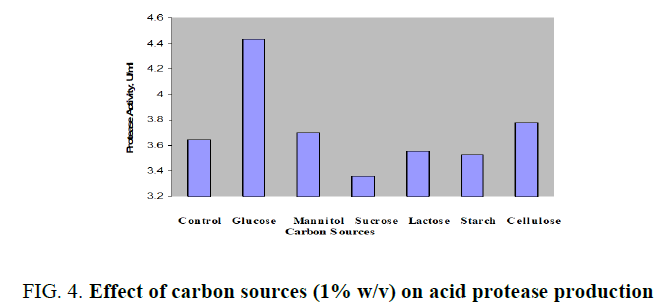
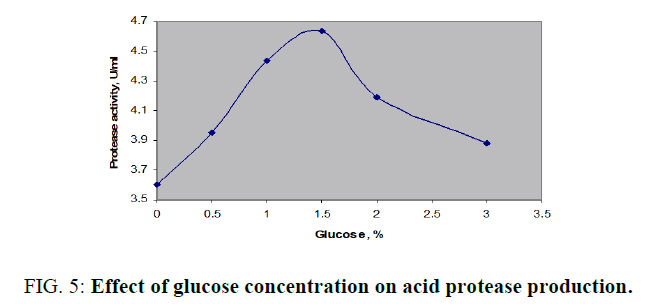
Production of acid protease with molasses as the control and supplementation of 1% (w/v) additional mono-, di-, and polysaccharides was displayed in FIG. 4. Peak enzyme activity was noticed with glucose as 4.438 U/ml. Slight increases in activity was found with mannitol or cellulose while the both tested disaccharides, sucrose and lactose were not significant additional carbon sources. Further optimization of glucose over an operating range up to 3% (w/v) was investigated and enhanced enzyme productivity of 4.638 U/ml was achieved with 1.5% (FIG. 5). Variation of enzyme activity with glucose concentration was clearly indicated by bell shaped curve. There was a continuous increase in the production up to 1.5% further it was dropped from 4.638 to 3.812 U/ml in the range of 1.6 to 3.0 % (w/v).
FIG. 4. Effect of carbon sources (1% w/v) on acid protease production
FIG. 5: Effect of glucose concentration on acid protease production.
Similar observation was noticed for acid protease production from fungal culture [8]. In addition, production media with multi carbon substrate source, brewery waste, was efficient in increased simultaneous production of amylases and proteases from Bacillus subtitles [13].
Partial purification of protease
The enzyme was harvested with filtration using Whatman No.1 and filtrate was collected. In the next stage of purification, enzyme from clarified liquid was obtained through another solid-liquid separation process, centrifugation. Diluted crude enzyme solution was concentrated by salt precipitation. Soluble protein mixture in the supernatant was converted to precipitated form with the addition of ammonium sulphate at 85% (w/v) saturation.
The precipitate obtained was dissolved in 0.05 M Tris-HCl buffer. A membrane separation process dialysis was employed for the removal of ammonium sulphate salt. Proteins were retained by membrane in retentate while the salt was appeared in permeate. Semi purification of acidic protease from various microorganisms was reported in earlier studies [7,14].
Conclusion
In this study, production medium of acid protease from Aspergillus spp. was designed through OVAT method with low cost substrates. Fungal culture was being capable of transforming sugar and dairy industrial effluents to extracellular acid protease. Enzyme activity was augmented from 3.648 to 4.638 U/ml with equal volumes of molasses and cheese whey, 1% (w/v) soya bean meal and 1.5% (w/v) of glucose at an optimal pH of 4.5 and five days of incubation. Protease was partially purified through precipitation by ammonium sulphate followed by dialysis with enhanced the activity of 5.545 μ/ml.
References
- Rao MB, Tanksale AM, Ghatge MS, et al. Molecular and biotechnological aspects of microbial proteases. Microbiology and molecular biology reviews. 1998; 62(3):597-35.
- Souza PM, Bittencourt ML, Caprara CC, et al. A biotechnology perspective of fungal proteases. Brazilian Journal of Microbiology. 2015;46(2):337-46.
- Djamel C, Ali T, Nelly C. Acid protease production by isolated species of Penicillium. European Journal of Scientific Research. 2009;25(3):469-77.
- Vishwanatha KS, Rao AA, Singh SA. Acid protease production by solid-state fermentation using Aspergillus oryzae MTCC 5341: Optimization of process parameters. Journal of industrial microbiology & biotechnology. 2010;37(2):129-38.
- Radha S, Nithya VJ, Babu RH, et al. Production and optimization of acid protease by Aspergillus spp under submerged fermentation. Archives of Applied Science Research. 2011;3(2):155-63.
- Shuler ML, Kargi F. A text book of Bioprocess Engineering Basic Concepts. 2003; 6(2):60-73.
- Xu Y, Dai M, Zang J, et al. Purification and Characterization of an Extracellular Acidic Protease of Pediococcus pentosaceus Isolated from Fermented Fish. Food Science and Technology Research. 2015;21(5):739-44.
- Sinha S, Sinha S. Studies on the production of acid protease by submerged fermentation. International journal of food engineering. 2009;5(1).
- Lowry OH, Rosebrough NJ, Farr AL, et al. Protein measurement with the Folin phenol reagent. Journal of biological chemistry. 1951;193(1):265-75.
- Benkahoul M, Benchiheub M, Bellil I, et al. Physical and chemical properties of the acid protease from Onopordum acanthium: Comparison between electrophoresis and HPLC of degradation casein profiles. African Journal of Biotechnology. 2016;15(9):331-340.
- Pant G, Prakash A, Pavani JV, et al. Production, optimization and partial purification of protease from Bacillus subtilis. Journal of Taibah University for Science. 2015;9(1):50-55.
- Souza PM, Werneck G, Aliakbarian B. Production, purification and characterization of an aspartic protease from Aspergillus foetidus. Food and Chemical Toxicology. 2017;109:1103-1110.
- Blanco AS, Durive OP, Pérez SB, et al. Simultaneous production of amylases and proteases by Bacillus subtilis in brewery wastes. brazilian journal of microbiology. 2016;47(3):665-674.
- e Silva TA, Knob A, Tremacoldi CR, et al. Purification and some properties of an extracellular acid protease from Aspergillusclavatus. World Journal of Microbiology and Biotechnology. 2011;27(11):2491-2497.