Original Article
, Volume: 12( 12)Multi-Drug Resistant Genes in Bacteria and 21st Century Problems Associated with Antibiotic Therapy
- *Correspondence:
- Asit Kumar Chakraborty , Department of Biotechnology & Biochemistry, Oriental Institute of Science & Technology (OIST), Vidyasagar University, Midnapore 721102, West Bengal, India, Tel: +919339609268; E-mail: chakraakc@gmail.com
Received: September 24, 2016; Accepted: October 13, 2016; Published: October 18, 2016
Citation: Asit KC. Multi-Drug Resistant Genes in Bacteria and 21st Century Problems Associated with Antibiotic Therapy. Biotechnol Ind J. 2016;12(12):114.
Abstract
Bacterial infections are cured by antibiotics since 1930s but recently such antibiotics would not work to cure most bacterial infections due to accumulation of many MDR genes in bacterial plasmids. Most notorious MDR gene is β-lactamase gene (bla) which hydrolyses lactam ring CO–N bond of penicillin. Other MDR genes include tet gene isomers which encode a membrane-bound drug efflux protein (~400 aa) which kicks out tetracycline from bacterial cell cytoplasm. strA/B gene encodes an enzyme (~267 aa) that phosphorylates streptomycin and phosphorylated streptomycin could not bind bacterial 50S ribosome. Similarly, diversified aad and aph MDR genes adenylate and phosphorylate aminoglycoside antibiotics which then could not able to bind ribosome to kill bacteria. cat gene acetylates chloramphenicol and acetylated chloramphenicol could not bind 30S ribosome Diversified aminoglycoside acetyl transferases (aacA1 and aacC1) also acetylate at various position of neomycin, amakacin and gentamycin. arr gene ribosylates refamycins which then could not inhibit bacterial RNA polymerase. Sul1/2 genes have been implicated in sulfamethoxazole resistant in the recent outbreaks of Stenotrophomonas maltophilia. VanA gene cluster are involved in the vancomycin resistance in Enterococcus facium as well as Staphyloocucus aureus and recently in Escherichia coli. ermA/B genes are diverged 23S rRNA methyl transferases that give resistant to macrolides. Other potential genes are MFS, RND and MATE types drug efflux genes that could kick out drugs in a proton-pump mechanism. Similarly, certain drug ABC transporters efflux antibiotics like doxorubicin with ATPase activity and mcr-1 gene (phosphoethanolamine transferase) gives resistant to colistin drug. Thus, we see that diverse MDR genes are present in E. coli, K. pneumoniae, S. aureus, S. enterica, P. aeruginosa, and many other common pathogens both located in large conjugative plasmids and chromosome. The author concluded that golden era of antibiotic had ended within 80 years of its discovery and alternative medicines like heterogeneous herbal antibiotics, bacteriophage therapy and gene medicines (antisense, ribozyme and miRNA) were now popular all over the world.
Keywords
Conjugative plasmids; mdr genes; Superbugs; Drug resistomes; Antibiotic therapy
Introduction
Multi-drug resistant bacteria hydrolyse the penicillin and cephalosporin drugs and recently few superbugs like KPC-2 Klebsiella pneumoniae and NDM-1 Escherichia coli hydrolyse potent carbapenem drugs as well as β-lactamase inhibitors like cavulinate and sulbactam [1-6]. Diversified Beta-lactamase enzymes (250 aa to 386 aa) in plasmids are the main cause of such drug resistance [7-10]. Since the discovery of penicillin in 1928 by Alexander Flaming from slime mold Penicillium notatum, numerous antibiotics like polymyxin, tetracycline, ciprofloxacin, streptomycin, neomycin, azithromycin and chloramphenicol had been discovered from actinomycetes or fungi. Such discoveries happened so successfully that we could destroy any pathogens by targeting cell wall, protein, DNA and RNA synthesis of bacteria without affecting human central dogma process [11,12]. However, discovery of antibiotic hydrolyzing enzymes (amp, tet, cat genes) as early as 1940s and their appearance in high copy number plasmids of common pathogens so instrumental that bacteria easily could grow in presence of 50 μg/ml of the drug in vitro [13-15]. Scientist warned the rapid increase of drug resistant bacteria in the environment and suggested to reduce antibiotic over dose in patients unless necessary and to reduce antibiotic use in animal growth and in agricultural land. Now a day more than thousands of MDR genes have been analyzed due to whole genome sequencing by colour di-deoxy method and easy analysis of large sequences by computer assisted programmes like BLAST [16,17].
The early beta-lactamase, penicillinase breaks the CO–N bond of benzyl-penicilin and penicillinic acid could not inhibit the peptidoglycan biosynthesis of bacteria. So, we got new derivatives, ampicillin and amoxicillin as potent drug that human used more than any drug to cure many disease epidemics since 1963. Then carbanicillin and methicillin were brought with success but altered penicillinase with more diverged sequences and broad spectrum β-lactamase activities were arose designated as CMY, OXA and AmpC β-lactamases. Presently, more than twenty different classes of bla enzymes have sequenced and millions of mutations are detected worldwide. In truth, more than 450 blaOXA isomers are detected and have no sequence similarities among OXA-1, OXA-23, OXA-24, OXA-48 and OXA-58.
However, the mechanisms of drug resistance are more grim than expected and have been divided into many fold apart from “bla resistome” as follows: a) drug efflux genes (acrAB, mexCD, mdt, tetA and norA; b) drug modifying genes like aminoglycoside acetyl transferases (aac), phospho transferases (aph) and adenyl transferases, (ANT) [18,19]; c) mecA gene and PBPs which catalyze the transpeptidase reaction even in the presence of high concentrations of β-lactam [20]; d) A lower expression of outer membrane proteins (OMPs) in imipenem resistance. The author had discussed current status on MDR genes, their discoveries, and occurrence and drug sensitivities in common pathogens.
Pre-Antibiotic Era, Antibiotic Golden Age and Re-Appearance of Antibiotic Dark Age
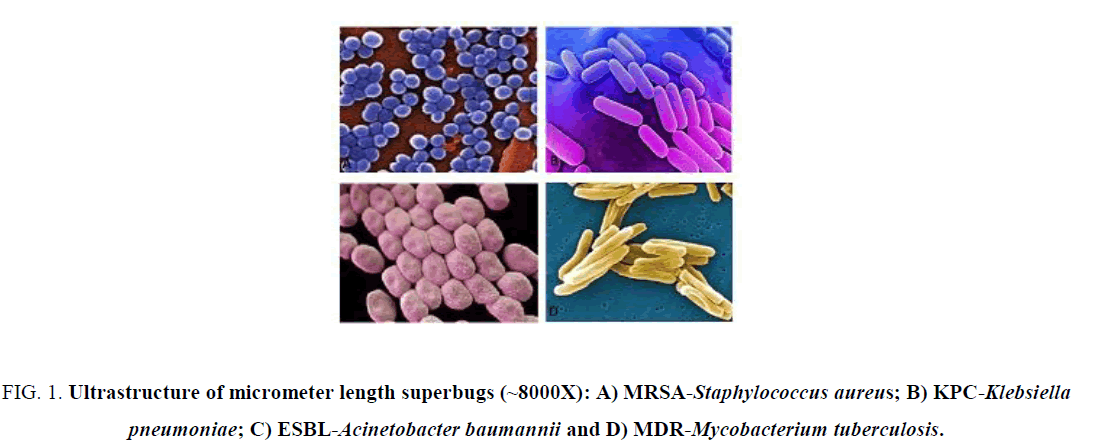
Benzyl penicillin was introduced to people in 1943. So, question arises how did then people save themselves from bacterial infections? Surely, 1000 plant extracts used to cure bacterial infections and sometime metal amalgam and wines were also used. Sanakrit books like Veda, Charaka Samhita and Sushruta Samhita described such medicinal plants. Presently, no antibiotic seems good to cure KPC2, NDM1 and OXA48- types bacterial infections as last resort drugs like imipenem, colistin and sulbactam resistant bacteria have emerged [9]. Question arises then if antibiotic dark age re-appeared again, even we have 10000 antibiotics in the selves? Sadly, drug companies also have slowed their new drug hunting due to MDR-void. During 1950-1990 was the golden era of antibiotics but uncontrolled increase of pollutions in 1960-1970 and aberrant use of antibiotics in crops and animal’s growth (1970-till date) had developed a gradual pick of mdr genes in bacteria [21]. Scientists warned repeatedly the MDR calamity as half of the common bacteria of the water were ampicillin, amoxicillin and tetracycline resistant. Now MDR bacteria were present in rain water, sea water and river water and everywhere and WHO suggested that at 2050, whole human and animal might face new MDR epidemics similar to past cholera, tuberculosis and pneumonia epidemics [22]. Major common MDR pathogens were, Pseudomonas aeruginosa, Streptococcus pneumoniae, Stenotrophomonas multophila, Micobacterium tuberculosis, Acinetobacter baumannii, Enterobacter aerogenes, Proteus mirabilis, Vibrio cholerae. Staphylococcus aureus and Escherichia coli and had drug resistant genes in plasmids as well as in chromosome in case of S. aureus and A. baumannii (Figure 1). Here, we will discuss the discovery of antibiotics and the respective MDR genes separately to demonstrate how such genes are amplified in conjugative plasmids (100 kb to 500 kb) and make common pathogens into superbugs.
Figure 1: Ultrastructure of micrometer length superbugs (~8000X): A) MRSA-Staphylococcus aureus; B) KPC-Klebsiella pneumoniae; C) ESBL-Acinetobacter baumannii and D) MDR-Mycobacterium tuberculosis.
Discovery of Penicillin and bla Genes
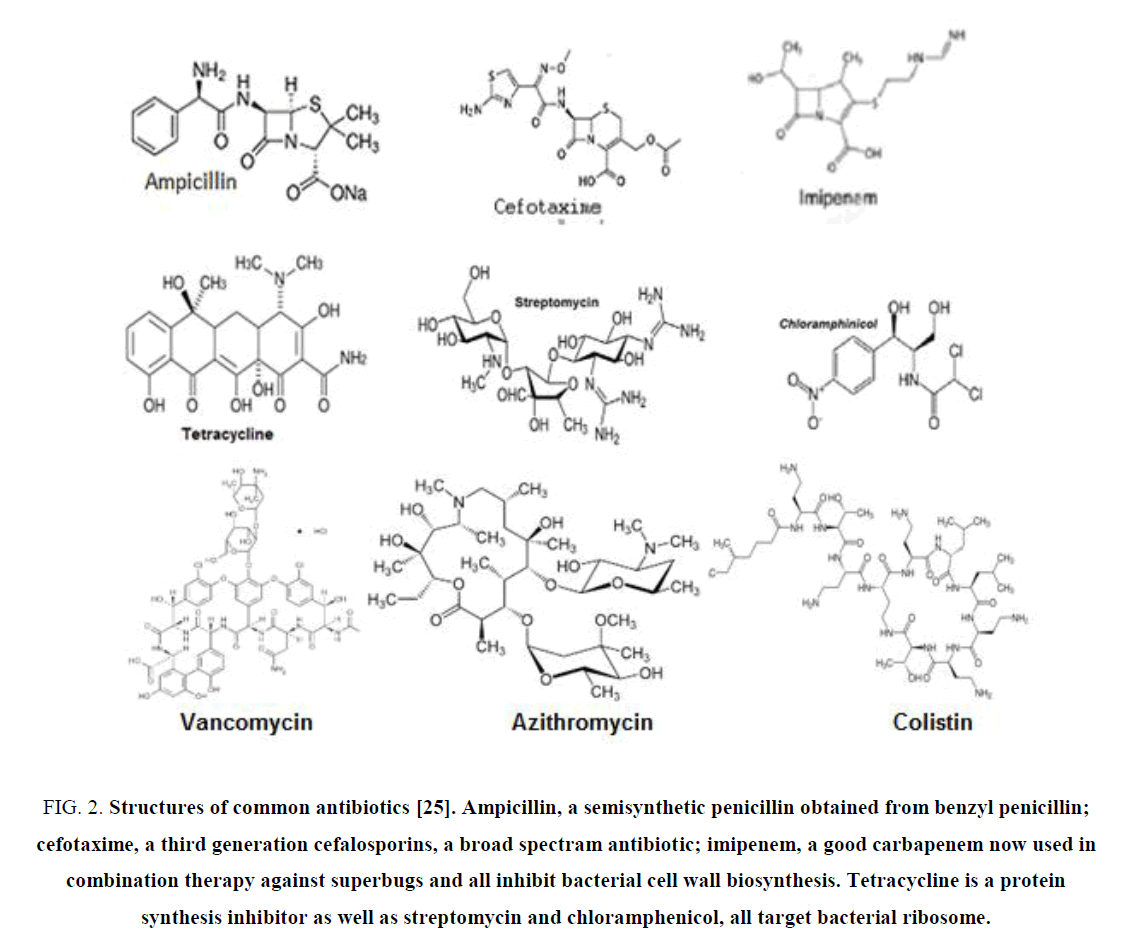
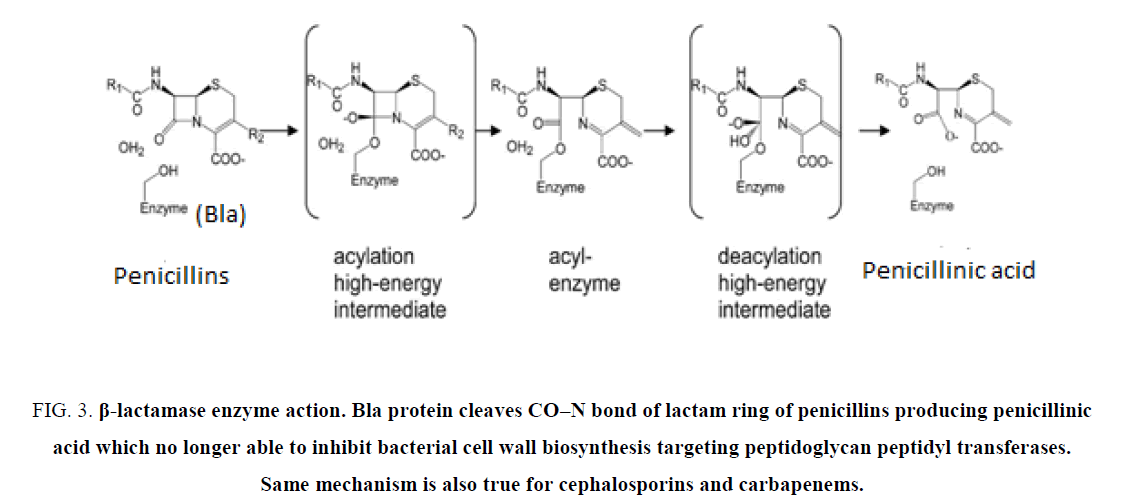
Penicillin first discovered in 1928 and it took until 1943 to commercialize benzyl penicillin for public use. But the penicillinase enzyme was discovered in 1940 that could break penicillin into useless penicillinic acid that could not stop bacterial peptidoglycan biosynthesis [23]. blaTEM-1 was first discovered in E. coli in 1965 from “Temora” named girl and blaSHV was mainly found in K. pneumonia was termed as “sulfhydryl reagent variable” [24]. Such β-lactamase genes mainly were found in IncA plasmids and integrons but other class bla enzymes were encoded on the chromosome like blaOXA-48 of A. baumannii or blaKPC of K. Pneumoniae. Thus, penicillin-G resistance due to class A PC enzyme, was overcame by introducing semisynthetic drugs like ampicillin, amoxycillin and then carbanicillin and oxacillin in 1960s. But soon class D OXA enzymes were involved in MDR-bacteria and again scientists synthesized new drugs, cephallosporins (Figure 2). Importantly, now many third generation cephalosporins like cefotaxime, ceftrioxane, and cefoxitin (1980) are used today in poor countries to get rid of bacterial infections. However, ESBL enzymes (blaKPC, blaSME, blaSHV180, OXA-210) could hydrolyse cephamycins and higher derivative like cefotaxime. Now most potent carbapenem drugs (imipenem, dorripenem, meropenem) were introduced as well as latest β-lactamase inhibitor (cavulinate, sulbactam, avibactam) combination therapy [25,26]. Sadly, carbapenemsaes named β-lactamases are spreading rapidly like KPC-16, VIM-10, OXA-48, OXA-450 and IMP-18 variants. The list of β-lactamase mutants in large conjugative plasmids of E. coli, K. pneumoniae, Haemophilus influenza, P. aeruginosa, S. aureus and Neisseria gonorrhoeae were detected in India, UK, China and USA. New Delhi metallo-β-lactamase (NDM-1) has been found in 2008 to confer Enterobacteriaceae with complete resistance to all β-lactam antibiotics including the carbapenems (Figure. 2). NDM-1-positive E. coli are now widespread in the environment and water supplies in India, Pakistan, Nepal because NDM-1 genes are located on conjugative plasmids that are prone to rearrangement and gene donation to non-MDR bacteria [27]. The mechanism of drug hydrolysis by all β-lactamase enzymes is simple, as penicillinic acid is an inactive product due to hydrolysis of CO–N bond of ring A of β-lactams (Figure 3).
Figure 2: Structures of common antibiotics [25]. Ampicillin, a semisynthetic penicillin obtained from benzyl penicillin; cefotaxime, a third generation cefalosporins, a broad spectram antibiotic; imipenem, a good carbapenem now used in combination therapy against superbugs and all inhibit bacterial cell wall biosynthesis. Tetracycline is a protein synthesis inhibitor as well as streptomycin and chloramphenicol, all target bacterial ribosome.
Figure 3: β-lactamase enzyme action. Bla protein cleaves CO?N bond of lactam ring of penicillins producing penicillinic acid which no longer able to inhibit bacterial cell wall biosynthesis targeting peptidoglycan peptidyl transferases. Same mechanism is also true for cephalosporins and carbapenems.
Discovery of Tetracycline and tet Genes
Tetracycline also was discovered at the end of 1940 when Streptomyces aureofaciens, Streptomyces rimosus, and Streptomyces viridofaciens were found to be excrete a novel compound with four ring structures (tetra) and the compounds were named as tetracyclins. Tetracycline chemistry is simple where four nucleus, A, B, C, and D rings are fused (Figure. 2). Nevertheless, a number of substitutions at different positions on the B, C, and D rings produced wonder drugs: doxycycline, minocycline and oxytetracycline. The tetracyclines are strong chelating agents and both their antimicrobial and pharmacokinetic properties are influenced by chelation with metal ions. It is well established that tetracyclines inhibit bacterial protein synthesis by preventing the association of aminoacyl-tRNA with the bacterial 50S ribosome. OmpF and OmpC porin channels are involved in tetracycline transport as positively charged cation with Mg+2 ion.
Similar to bla genes that confer resistant to ampicillin antibiotic, tetracycline resistant is mediated by different method with ~400 amino acids Tet protein which belongs to major facilitator superfamily (MFS) similar to drug efflux protein acrAB or mexCD (Table 1). Export of tetracycline reduces the intracellular drug concentration and thus protects the ribosomes within the cell. Thus, Tet protein does not alter tetracycline structure like Bla protein does alter ampicillin structure. Tet efflux genes isomers are found in both gram-positive and gram-negative bacteria and there are significant sequence variations as in tetA, tetB, tetC, tetD, tetG, tetE, tetS, tetT, tetK, tetL and tetT [28]. Such genes were cloned from E. coli, Shigella sp., Enterococcus sp., Streptococcus sp., Mycobacterium sp., Salmonella enterica and Vibrio cholerae. Minocycline and glycyclines are not always substrate for Tet protein and are good antibiotic to use in case of tetracycline resistance [29].
There are few ribosomal protection proteins that confer resistance to tetracycline in a different mechanism involving tet(M), tet(O), tet(S), tet(W) tet(Q), tet(T) otr(A), tet(P) etc. Similarly, the tet(X) gene encodes 44 KD enzyme, the only example of tetracycline resistance due to enzymatic alteration of tetracycline in the presence of both oxygen and NADPH. So, three different specific mechanisms of tetracycline resistance have been discussed: 1) tetracycline efflux; 2) ribosome protection and 3) tetracycline modification there by made wonder tetracycline drugs useless today. The MFS Tet export protein acts as an electro-neutral antiport system which catalyzes the exchange of tetracycline divalent metal cation complex for a proton. In gram-negative bacteria, the export protein contains 12 TMS whereas in gram-positive bacteria it displays 14 TMS. Ribosome protection is mediated by a soluble ribosome-binding protein which binds tetracycline with greater efficiency so that tetracycline no longer binds to ribosome. The third mechanism involves a cytoplasmic protein that chemically modifies tetracycline in the presence of oxygen and NADPH. The two first mechanisms are the most widespread and most of their genes are normally acquired via transferable plasmids and/or transposons [30].
Discovery of Streptomycin and strA/strB Genes
As the story of penicillins unfolds followed by cephalosporins, there were many other antibiotics were discovered as early as 1940s like tetracyclins, chloramphenicol and streptomycins, all three did inhibit the protein synthesis in bacteria. The streptomycin antibiotic is very famous in curing tuberculosis patient and called aminoglycoside antibiotic. It was discovered in 1942 by Selman A Walksman from Actinomycetes (soil bacteria named Streptomyces griseous) and obtained Nobel prize in 1952 for curing tuberculosis and penicillin or sulfa-drug resistant pathogens. Streptomycin chemistry is simple as on hydrolysis it produces streptidine, streptrose and N-methyl-glucosamine. It kills bacteria both gram (+) and gram (-) at 1 μg/ml and protect rat or mice at 100 μg/ml from Salmonella sp., Shigella sp., Brucella abortus, Pseudomonas aeruginosa, Klebsiella pneumoniae, Diplococcus pneumoniae, and a number of organisms commonly found in urinary tract infections. However, bacteria this time also managed to develop a new gene called “strA and strB” that inactivate the antibiotic by phosphorylating the drug and such bacteria could grow at as high as 1000 μg/ml streptomycin (Table 1) [31].
| Major Classification of Different MDR Genes in bacteria | ||||
|---|---|---|---|---|
| Genes | Full name of subclass | Amino acids | Accession number | Protein Id number |
| bla | blaTEM | 286 aa | J01749, X54606 | AAB59737, ABI20744 |
| blaCTX-M | 291 aa | X92506, X92507 | ABN09669, CAA63262 | |
| blaSHV | 286 aa | X98098, AF148850 | AAD37412, ABC25482 | |
| blaKPC | 293 aa | AF297554, HM066995 | AAG13410, ACB71165 | |
| blaNDM1 | 270 aa | KC539430, JF703135 | AGC54622, AFQ31613 | |
| blaOXA1 | 276 aa | AF227505, GQ896557 | AFG30109, CAC82805 | |
| blaIMP | 246 aa | S71932, EU352796 | AAB30289, AAG17724 | |
| blaVIM | 266 aa | AJ291609, AY987853 | ABV21756, CAC82722 | |
| blaAMP-C | 382 aa | AF124204, DQ092429 | AAD28044, AHN62490 | |
| blaSPM | 276 aa | AY341249, KC710242 | AAR15341, CAD37801 | |
| tet | tetA | 399 aa | X75761, KT950741 | CAA53389, ALS39162 |
| tetA | 424 aa | HM453327, KF705205 | AHC55487 | |
| tetB | 401 aa | KP899806 | AKJ20239 | |
| tetC | 396 aa | KC590080 | AGL61405 | |
| tetD | 394 aa | AB089602 | BAC67150 | |
| tetE | 405 aa | JN315882 | AEW70668 | |
| str | strA | 267 aa | M28829, KT225462 | AAA26443, ALF35537 |
| strB | 278 aa | NC_018107, LN555650 | CED95339, AFU91634 | |
| cat | catB3 | 210 aa | EF516991, DQ343904 | ABP52023, ABC69169 |
| aac | aacA1 | 185 aa | AB061794, AB901039 | BAB72153, BAO48019 |
| aacA4 | 152 aa | JN596279, JX486753 | AEZ05102, AFV31448 | |
| aac(3’) | 286 aa | M62833 | AAA21890 | |
| aac(2’) | 210 aa | U72743 | AAB41701 | |
| aad | aadA1 | 263 aa | AF324464, DQ522230 | AAK13440, ABG23480 |
| aadA4 | 263 aa | AY138986 | AAV34365 | |
| aadA5 | 262 aa | KT175895 | ALO62079 | |
| aadA7 | 265 aa | DQ520937 | ABG01709 | |
| aph | aph(6’) | 266 aa | X01702, Y00459 | CAA25854, CAA68516 |
| aph(4’) | 341 aa | V01499, X03615 | CAA24743, CAA27276 | |
| aph(3’) | 264 aa | U32991, V00618 | AAA85506, CAA23892 | |
| aph(2’) | 294 aa | NC_018107 | WP_000155092 | |
| RND | acrAB/EF | 1027aa | M94249, V00734 | WP_001132469 |
| mexAB/CD | 1045aa | L11616, U57969 | WP_023101049 | |
| MFS | norA | 388 aa | D90119 | BAA14147 |
| mdtE | 385 aa | CP000247 | ABG71588 | |
| ABC | macB | 664 aa | APBE01000048 | EMU20415 |
| DrrA | 316 aa | HE971709 | CCK28451 | |
| SMR | Mmr | 107 aa | LHCK01000002 | KPU48951 |
| emrE | 110 aa | Z11877 | CAA77936 | |
| EMR | emrA/B | ~243aa | X03216, Y00116 | CAA26964, CAA68299 |
| MCR | mcr-1 | 541aa | NG_050417 | WP_049589868 |
| VanHAX | vanA | 287aa | KR047792 | AKE81063 |
| DHPS | sul1/2 | ~279aa | KM877269, AP012056 | AKG90139, BAN19562 |
Table 1: Major classification of MDR genes that are located mostly in plasmids but also in bacterial chromosome. Those genes inactivate the antibiotics by diverse mechanisms and such bacterial infections are hard to treat with common antibiotics. MDR genes are activated by IS-elements, transposons and mobile elements but also are integrated and diversified in F-plasmids that has facilitated spread of MDR genes into common pathogens.
Further, other MDR genes that were found in many conjugative MDR plasmids were aph and aad genes that conferred bacteria resistant to aminoglycoside antibiotics. The aminoglycoside phosphotransferase (aph) phosphorylate the antibiotics so that phosphorylated kanamycin, amikacin and neomycin could not kill the bacteria. Antibiotic Adenyl transferase (aad) seems Rel-Spo-like super family and does not has much similarity to the AG 3’ N-acetyl transferases and AG phospho transferases. This enzyme has similarity across the species and notably exists as different isomers with 50% to 80% sequence similarities. The enzyme has divided into aadA1 to aad17 according to mutation and sequence similarities. The aadA1 is usually 263 amino acids long and can adenylate variety of aminoglycosides at the 6-N position as in streptomycin and amikacin. The aminoglycoside adenyl transferase [EC:2.7.7.47] was present in many bacterial plasmids of diverse species of Escherichia (accession nos. HG41719, KJ484637, KM377239), Klebsiella (accession nos. KF914286, KF719970), Salmonella (accession no. JQ899055), Acinetobacter (accession no. KM401411), but also present in some bacterial chromosome as in Salmonella enterica [32].
Discovery of Chloramphenicol and cat Gene
Chloramphenicol was first discovered in actinomycetes isolated from soil by Ehrlich in 1947. Chloramphenicol inhibits the bacterial 30S ribosome and is a very good antibiotic as it cures the common diseases caused by S. aureus, E. coli, K. pneumoniae (nosocomal disturbances), and more. Chloramphenicol was also isolated from Streptosporangium viridogriseum var. kofuense by Tamura in 1971 and from the marine snail Lunatia heros by Price in 1981. However, soon cat gene was discovered in many bacterial pathogens those were found resistant to chloramphenicol. Cat enzyme is different from “amp” or “tet” gene’s mode of actions in that it acetylates the chloramphenicol drug in such a way that acetylated drug no longer able to bind the bacterial ribosome [33]. Thus, cat gene containing bacteria (in plasmid) easily grow in media containing MIC amount of chloramphenicol in vitro as well as in vivo in patient blood. (Figure 4). cat gene is structurally different to aac gene that also acetylates aminoglycoside antibiotics.
Figure 4: Detection of cat gene activity which acetylates chloramphenicol [34]. cat gene was cloned in plasmid pBL cat and transfected into HeLa cells, G418 selected and cells extracts were assayed for cat enzyme activity and TLC separated. Lane 1, Fluorescen-labelled chloramphenicol, lane 2 + normal HeLa cells extract, lane 3 + 5 μl G418 selected extract, lane 4 + 10 μl G418 selected extract. It was shown that acetylated chloramphenicol moved fast in TLC gel in accent chromatography in organic media.
Discovery of Fluoroquinolones and qnr Genes
Oxolinic acid and nalidixic acid was the first drug and ciprofloxacin was the mostly used derivative. Bayer discovered ciprofloxacin in 1981 and FDA approved first in the USA in 1987. Ciprofloxacin was mostly used in UTI and lower respiratory infections (pneumonia, bronchitis). Fluroquinolones are famous drug with hundred derivatives including norfloxacin, enoxacin, ofloxacin, moxifloxacin, gatifloxacin, levofloxacin and gemofloxacin. Fluoroquinolones are bacterial type II topoisomerase (DNA gyrase) inhibitors that affect the DNA replication (DNA duplication during cell division) as well as transcription (RNA synthesis from DNA i.e. mRNA, tRNA and rRNA) and transposition (movement of DNA segment by duplication and integration into a new position involving integrase, transposase, and resolvase enzymes). In other word, gyrase inhibitors control DNA supercoiling and thus bacterial cell division stops. Fluoroquinolones are the improved derivatives used now a day but drug resistant bacteria against those drugs are evident now due to mutations in the gyrA and gyrB subunits of DNA gyrase enzyme demonstrating another kind of drug resistant mechanism by altering the target site itself [35]. norA/B efflux protein also involved in some bacteria that are resistant to fluoroquinolones. Qnr protein binds to fluoroquinolone and make the drugs ineffective to kill bacteria. Thus, we found many gene creations in bacteria to make it drug resistant (Table 1) [36].
Discovery of Macrolides and erm Genes
Erythromycin, the first macrolide antibiotic, was used since 1950s for the treatment of upper respiratory tract and skin and soft tissue infections caused by many microorganisms including Legionella, Mycoplasma, and Chlamydia, especially in the penicillin-allergic patient. In 1991, the US Food and Drug Administration (FDA) approved clarithromycin and azithromycin for the treatment of sexually transmitted diseases, respiratory tract infections and infections caused by Helicobacter pyroli that causes intestinal cancer as well as Mycobacterium avium complex that causes tuberculosis. During the last few decades, erythromycin resistance has been reported everywhere mainly in Streptococcus pyogenes and Streptococcus pneumoniae. However, azithromycin resistance has reached now in household bacteria like E. coli. Macrolide drug resistance is mediated by mainly erm class of genes that code methylases which spefically methylate 23S rRNA. Post-transcriptional methylation of an adenine residue in 23S rRNA results in co-resistance to macrolide, lincosamide, and streptogramin B antibiotics (MLS phenotype). ermA and ermB DNA methylase are 243 aa and 245 aa long respectively. Further efflux-mediated erythromycin resistance is common in streptococci for 14- and 15- membered macrolides (M phenotype). Active efflux is encoded by mef-class genes (discovered first in S. pyogenes) that were subsequently found to be widespread also in S. pneumoniae and another streptococcal species. A closely related mel gene (msrA homolog), encoding a proton motive force pump and a putative ATP-binding cassette transporter homolog actively effluxs erythromycin in a proton-motive force [37]. Thus, target site alteration and drug efflux mediated multiple MDR mechanisms exists also in azithromycin resistance.
Discovery of Sulfonamides and sul1/2 Genes
Sulphonamide is first synthetic pro-drug marketed by Bayer in 1935 and successfully is used during world war II saving the lives of tens of thousands of patients, including Franklin Roosevelt, Jr. and Winston Churchill. Sulfonamides are structural analogues of para amino benzoic acid (PABA), which competitively inhibit dihydropteroate synthetase activity blocking folic acid synthesis. Combination of trimethoprim (dihydrofolate reductase inhibitor) and sulfamethoxazole with the trade name of cotrimoxazole is the first antibiotic that has been used for the treatment of urinary tract infections [38]. Sul1 and sul2 genes encode altered dihydropteroate synthese that found mostly in class 1 integron plasmids and many MDR plasmids conferring sulphonamide and sulphamethoxazole resistance. In particular, Salmonella enterica serotype typhimurium definitive phage type 104 with resistance to ampicillin, streptomycin, sulfonamides, chloramphenicol and tetracyclines has emerged as a global health problem. Sul1/2 genes were found in many MDR plasmids and another Sul3 gene also detected in many plasmids that differs in DNA sequence [39].
Discovery of Polymyxins and mcr-1 Gene
Polymyxins are heterogenous with respect to amino acid chains and fatty acid side chains and classified as polymyxin- A, -B, -C, -D, and -E. The polymyxin B and polymyxin E are very famous as colistin is now used in penicillin and tetracycline resistant pathogenesis. The main difference between these two molecules is that polymyxin B contains phenylalanine in position 6, while colistin contains D-leucine. Polymyxin B was first isolated from Paenibacillus polymyxa. Its tripeptide side chain is linked to the fatty acid residue, which has been identified as 6-methyl-octan-oic acid (polymyxin B1) or 6-methyl-eptanoic acid (polymyxin B2). Polymyxins bind to the gram-negative bacterial cell membrane phospholipids, producing a disruptive physicochemical effect, which leads to cell-membrane leakage and cell death [40]. A secondary target site for polymyxins is the type II NADH-quinone oxydoreductase respiratory enzyme (electron transport pathway) of the inner bacterial membrane of both gram (+) and gram (-) bacteria. In November 2015, Liu et al. [41] reported first the plasmid-mediated colistin resistance gene mcr-1 in E. coli isolates from animal, food and patients in China. Now it spreads worldwide like USA and UK similar to superbug gene blaNDM1 that confers resistance to carbapenems. mcr-1 codes an enzyme called ethanolamine transferase that transfer ethanolamine to lipid A. Colistin use is restricted in Europe in veterinary medicine to oral use for the treatment of enteric diseases (750000I U/kg body wt/day/7days) including China but at reduced dose (1/4th of that in Europe). Because colistin poorly absorbed in the intestine, low dose has facilitated >30% resistant GI bacteria to colistin in China diarrhoeal pigs.
Discovery of Vancomycin and VanA gene Cluster
Vancomycin largely used in penicillin resistance and subsequent streptomycin resistance against many Enterobacteriaceae. Such glycopeptides inhibit cell wall synthesis in bacteria by binding the C-terminal D-Ala-D-Ala of the pentapeptide precursors of peptidoglycan, thus blocking the transpeptidation and transglycosylation [42]. High level vancomycin resistance was 1st reported in 1988 after its 30 years use in clinical practice. Today seven types of vancomycin resistance genes were identified (VanA, B, C, D, E, G, and L). Such gene cluster changes D-Ala-D-Ala to D-Ala-D-Lactate (VanA, VanB, VanD mediated) or D-Ala-D-Serine (VanC/E/G/L mediated), thus continuing peptidoglycan synthesis in presence of vancomycin. VanA type resistance is common in MRSA S. aureus isolates. VanH is a dehydrogenase which is responsible for reducing pyruvate into lactate and VanA ligates lactate into D-Ala-D-Lactate which acts as precursor for pentapeptide that is involved in transpeptidase reaction to make cell wall peptidoglycan (cross linking among muraminic acid pentapeptides through (Gly)5 bridge). A 11 kb VanA operon also contains VanX/Y dipeptidase which removes D-Ala-D-Ala from system by hydrolysis and VanR/VanS which are transcriptional regulator (two component sensor system) of the VanA operon where VanR is activator of VanA/H/X/Y. On the other hand, VanS regulates VanR by phosphorylation and in presence of vancomycin. VanS is highly phosphorylated at the histidine residue for its activation. Our recent data suggested that MDR E. coli of Kolkata is highly resistant to vancomycin but molecular VanA type has not detected yet.
Wide-Spread Multi-Drug Resistant Bacteria and Mechanisms of Drug Resistance
It is evident that any stress creates new genes in bacteria to alleviate toxicity. Thus, taking excessive prescription drug during bacterial infection creates drug resistant bacteria and during continuous exposure of many antibiotics, such bacteria acquires multiple genes for drug cleavage or modification or export. Those genes are known as mdr genes and we have had discovered gradually hundreds of such genes (Table 1) from patient’s blood, urine, pus etc. during the treatment regime of human population across the world. It is evident that drug resistant genes may present in nature due to cross selectivity and survival of multiple organisms like bacteria, actinomycetes, fungi and algae in nature. But the best selectivity and identification during treatment of patients have been established (in the patient’s gastrointestinal track and in blood in case of sepsis) in scientific community today. Interestingly, healthcare-associated infections have occurred in the hospitals are main cause of multi-drug resistant bacterial spread worldwide. We can classify four broad areas that cause untreatable blood infections in human populations that we must know: 1) Catheter-associated bloodstream infections; 2) Ventilator-associated pneumonia; 3) Surgical site infections of Acinetobacter baumannii and 4) Catheter-associated urinary tract infections. However, as the MDR bacteria present in sea and river water, in air dust, and in beds, books and other households, any one may get infection any time if one’s immune system weak [4].
E. coli bacteria normally lives in the intestine of human and discovered as early as in 1885 by German bacteriologist Theodor Escherich. There are hundreds of types of this gram (-), facultative anaerobe rod-shaped bacteria and may be harmless even some are beneficial as produce important vitamins, such as vitamin K and B-complex. Specially, strains O104:H4 and O157:H7 of E. coli are very harmful to human as secrete potent Shigilla toxins causing diarrhea. E. coli belonging to the H30-R strain and H30-Rx are very resistant to fluoroquinolones the drug used to treat UTI. It was discovered recently further mutations in U30-Rx termed generation of E. coli superbug strain ST131 family as a major cause of urinary tract and kidney infections as well as sepsis in women, elderly and children. The ST131 bacteria were notable because they had acquired resistance to a many class of fluoroquinolones and acquired genes for extended-spectrum beta-lactamase causing resistant to new drug, carbapenems. Types of CRE are sometimes known as KPC (Klebsiella pneumoniae carbapenemase) and NDM (New Delhi Metallo-beta-lactamase). KPC and NDM are enzymes that break down carbapenems and make them ineffective and present in associated with other mdr genes in large plasmids (Table 2).
| Multi-drug resistant (mdr) genes in bacterial conjugative large Plasmids | ||||
|---|---|---|---|---|
| Accession number | Size (kb) | Beta-lactamases, drug transporters, metal resistant and antibiotic inactivating enzymes were found | GenBank Year | MDR Pathogens |
| NC_018107 | 353 | emrE, nhaA, crcB, NAT, terA/D, aph*, blaTEM, | 2015 | K. oxytoca |
| NC_022078 | 317 | ABC, merB, cat, aph*, aac3’, cmr, tetA, blaKPC, | 2015 | K.pneumoniae |
| LN555650 | 299 | terF, sul1, strA, catB, blaACC-1, aacA4, blaVIM-1 | 2015 | S. enterica |
| KM877269 | 249 | aad, floR, hph, aac6’-1b-cr, blaOXA, catB, arr3, sul1 | 2015 | S. enterica |
| CP011634 | 227 | blaOXA, aad, blaTEM, merC, aad, sul1, aac, blaTEM | 2015 | K. oxytoca |
| HG530658 | 223 | terW, blaACC-1, strA, aadA2, aac3’, rcnA, pcoS | 2015 | E. coli |
| NC_021087 | 26 | blaGIM-1, aacA4, aadA1, blaOXA-2, sul1 | 2015 | E. cloacae |
| LN850163 | 167 | MFS, AAA tetA, cat, blaTEM, macB, blaCTXm | 2015 | E. coli |
| KT185451 | 151 | blaTEM/CTXm/SHV12/KPC, merD, blaNDM1 | 2015 | K. pneumoniae |
| KF705205 | 134 | hph, strA, aac(3’)-IV, tetA, blaTEM-1 | 2015 | S. enterica |
| KP893385 | 137 | blaCTXm-65, blaKPC-2, blaSHV-12, blaTEM-1b | 2015 | K. pneumoniae |
| KT225462 | 50 | mphE, mare, sul1, blaDHA-1, qbrB4, strA, strB | 2015 | K. pneumoniae |
| LC055503 | 160 | blaSHV12, aac6’, blaOXA10, aadA1, sul1, blaDHA1 | 2015 | K. pneumoniae |
| KC543497 | 501 | Ter2, blaOXA-10, MFS, blaTEM8, ble, catB8, aac | 2014 | P. aeruginosa |
| NC_012690 | 148 | floR, tetA, strB, sul2, blaAmpC, sul1, aph , blaTEM1, | 2014 | E. coli |
| KF954760 | 140 | blaTEM1, strA, strB, NcrC, YihA, aadA | 2014 | K. pneumoniae |
| HG941719 | 135 | blaTEM1, aadA5, mphA, blaCTX, blaOXA, aac6, sulI, tetA | 2014 | E. coli |
| NC_020087 | 133 | aphA, hph, tetA, blaLAP2, dhfrXII, ble, qnrS1 | 2014 | K. pneumoniae |
| CP009115 | 118 | ble, blaOXO, qnr, ble, MerA | 2014 | K. pneumoniae |
| NC_019375 | 180 | blaVIM, aacA7, dhfr, ANT3’, SHV-5, sul1, aph3’ | 2014 | P. stuartii |
| NC_022522 | 168 | blaCTX-M25, aacA4*, strB, strA, aadB, blaOXA21 | 2014 | S. enterica |
| NC_012692 | 167 | strA, blaCMY2, groEL, stbA, strB, floR, merA | 2014 | E. coli |
| NC_019121 | 166 | blaAmpC, sul2, tetA, floR, TniB, mcp, hygBR, aph | 2014 | S. enterica |
| CP007558 | 272 | blaAmpC, ABC, sul1, blaTEM, aad, ble | 2014 | C. freundii |
| CP009116 | 95 | Aph, blaTEM, aac3’, MFS, dhfr, aad, arr2, blaNDM1 | 2014 | K. pneumoniae |
| NC_019889 | 87 | Aac(3’)-II, blaNDM-1, sul1, MsrE, mphE | 2014 | K. pneumoniae |
| NC_024978 | 110 | dhfr, aad3, blaCTX1, EtBrR, ABC-type | 2014 | E. coli |
| KJ541071 | 44 | sul1, blaOXA-2, aadA/B, blaTEM, catA1, blaGES-5 | 2014 | E. coli |
| KF954759 | 73 | blaKPC3, strB, aac(6’), chrB, ncrA/Y, srbA | 2014 | K. pneumoniae |
| AP012055 | 250 | blaNDM1, ccdA, ccdB, aadA2, catA1, qacA1 | 2013 | K. pneumoniae |
| AP012056 | 141 | Aac3/6, catB4, tetA, sul2, blaOXA/CTX/TEM, strB/A | 2013 | K. pneumoniae |
| JX566770 | 107 | pac, aadA1, dhfrA1, strB, blaTEM, ydiA | 2013 | E. aerogenes |
| JX442976 | 172 | tetA, aph, sul2, aadA1, blaOXA-10,Qnr, blaCMY-16 | 2013 | K. pneumoniae |
| JX104759 | 42 | blaKPC2, ABC, pliX3 | 2013 | K. pneumoniae |
| KC354802 | 41 | aacA4, aadA1, blaOXA-9, TEM-1 | 2013 | K. pneumoniae |
| JN420336 | 267 | blaNDM1, blaOXa1, aac6’, qnrB1, cat, blaCTX-M, | 2012 | K. pneumoniae |
| JX283456 | 108 | blaKPC2, TolA, blaTEM, ABC transporter, mph2, | 2012 | K. pneumoniae |
| AY046276 | 51 | aadA1, blaOXA-2, sul1, tetA, arsA/C, ABC | 2012 | S. enterica |
| AB61665 | 47 | blaIMP2, aacA4, aadA2, tetA, blaCTXm, sul1 | 2012 | K. pneumoniae |
| JN215524 | 24 | Dhfr, cmlA, blaOXA10, aadA1, qnrB4, blaDHA1,sul1 | 2012 | C. freundii |
| GU256641 | 110 | Sul2, strA, blaTEM1, blaSCO1, aacC2, blaACC-4 | 2011 | E. coli |
| GU585907 | 79 | aadA2, aphA2, aadA1*, strA, strB, blaVIM1, merA | 2010 | K.pneumoniae |
| FJ628167 | 151 | blaKPC, sul1, qnrB4, blaDHA, merE, mph2, ABC | 2010 | K. pneumoniae |
Table 2: Accumulation of mdr-genes in large plasmids of MDR-bacteria. Superbugs plasmids that contains 10-15 drug resistant genes are shown here. Many of them contains multiple beta-lactamse genes, acetyl or phospho transferases and MFS drug efflux genes surrounded by transposons and Tra conjugative genes.
Both enzymes, as well as the enzyme VIM (Verona Integron-Mediated Metallo-β-lactamase) have also been reported in Pseudomonas. E. coli ST131, causes urinary bladder infections and for more than 3000 deaths a year. Enterobacteriaceae bacteria like Citrobacter freundii, Enterobacter aerogenes, Enterobacter cloacae, Morganella morganii, Proteus mirabilis, Klebsiella pneumoniae, Klebsiella oxytoca, Providencia stuartii apart from E. coli were shown to acquire varying degree of drug resistant involving tetracycline, ampicillin, chloramphenicol, streptomycin, cefoxitin, cefotaxime, ceftazidine and sometime most wonder drugs like meropenem, imipenem and doropenem (Table 2) [43]. MRSA bacteria is initially discovered as Methicillin resistant microbe named Staphylococcus aureus - a gram positive circular bacteria that notoriously known as for skin infections (Figure. 1). MRSA means it is not only resistant to methicillin but also resistant to diverse groups of antibiotics. It may also be activated by drug-resistant genes in plasmids and may such genes duplicated in its single genomic DNA. MRSA skin and wound infections is now a problem in hospitals worldwide and is increasingly spreading in community health centres.
The methicillin resistance gene (mecA) encodes a methicillin-resistant penicillin-binding protein and activated with mobile genetic element, the staphylococcal cassette chromosome mec (SCCmec), of many MRSA isolates which also associated with bla, aac, aad, ANT and Sul1/2 types MDR genes. Such multi-drug resistant bacteria are susceptible only to glycopeptides antibiotics such as vancomycin and tigicycline. However, we found highly vancomycin resistant E. coli in Kolkata streets MDR-bacteria. The first VanA-mediated, vancomycin-resistant Staphylococcus aureus (VRSA) strain was isolated in USA in the year 2002. VanA plasmid (57.9 kb) consisted of a pSK41-like S. aureus plasmid with an insertion of a Tn1546-like element carrying the VanA operon possibly originated from vancomycin-resistant Enterococcus (VRE). The VanA plasmid from this E. faecalis isolate was characterized by Flannagan et al. [44] having Inc18 plamids like pIP501 and pAM_1 [45].
K. pneumoniae carbapenemase (KPC)-producing MDR-bacteria causes infections associated with significant morbidity and mortality. In 2001, Yigit et al. [46] first described KPC-producing K. pneumoniae isolate which encoded blaKPC activated by Tn3-type transposon, Tn4401 (Table 2). This transposon is a genetic element which is capable of inserting into diverse plasmids of gram-negative bacteria. Plasmids carrying blaKPC are often also associated with resistance determinants for other antibiotics like streptomycin, tetracycline and Sul1/2 gene that is involved in sulfa-drug resistance [47]. blaKPC was often associated with 2-3 other bla gene isomers and drug modifying genes like aminoglycoside phospho transferases and such plasmids are widespread among many gram-negative bacilli like E. coli, Enterobacter cloacae, Salmonella enterica, Proteus mirabilis and Citrobacter freundii. Tigecycline is a novel antibiotic that is used in treatment of ESBL and XDR infections caused by KPC-producing bacteria and other multidrug-resistant gram-negative bacteria.
Acinetobacter is an aerobic, gram-negative, non-flagellated bacterium (Greek word means motionless) commonly found in soil and water while Acinetobacter baumannii are found in 80% of reported hospital infections. It is catalase positive but oxidase and indole test negative. Acinetobacter can enter through open wounds causing fever, chills, cough, meningitis, and urinary tract infection. Its remarkable ability to develop or acquire multiple antibiotic resistance and propensity to survive for prolonged periods under a wide range of environmental conditions, make it a frequent cause of hospital outbreaks and an endemic healthcare associated pathogen. A. baumannii resistant to antimicrobial drugs like aminopenicillins, second-generation cephalosporins, most aminoglycosides, chloramphenicol and tetracycline are frequently observed now and strains of A. baumannii have also gained resistance to newly developed 4th generation antimicrobial drugs like meropenem. Studies showed that A. baumannii nosocomal infections in several hospitals worldwide very persistent as resistant to all hundred β-lactams, including carbapenems. Such bacteria also express chromosomally located ESBL PER-7, OXA-23 and ArmA 16S rRNA methylase (PER-7 differs from PER-1 by 4 amino acid substitutions at Q119E, V245I, R246K, and A294T) [48-51].
Multidrug resistance in P. aeruginosa is due to diverse resistance determinants (blaAmpC, aac, sul1/2, strA/B, ANT, cat, tet, mdt) in plasmids flanked by transposons and IS-elements. Such plasmids are donating drug resistomes to common pathogens due to presence of type-III and type-IV secretary systems like Tra and Tpn genes. The resistance-nodulation-cell division (RND) family of drug transporters is responsible for clinically relevant drug resistance among gram-negative bacteria and facilitates active efflux of multiple antimicrobial substrates like doxacycline and imipenem including biocides, detergents and dyes. Such genes are also chromosomally located and are over-expressed following mutations in their respective negative regulator genes at the promoter-enhancer. At least seven RND efflux systems (e.g., MexAB-OprM, MexCD-OprJ, MexEF-OprN and MexXY-OprM) have been described in P. aeruginosa . The MexAB-OprM system has the broadest substrate specificity and contributes to resistance to macrolides, aminoglycosides, sulfonamides, fluoroquinolones, tetracyclines and many β-lactams. The loss of the outer membrane protein (porin) OprD, is associated with imipenem resistance and reduced susceptibility to meropenem. Expression of aminoglycoside modifying enzymes (acetyl transferases, nucleotidyl transferases and phospho transferases), mediating aminoglycoside resistance are common. Methylation of specific nucleotides within the A-site of 16S rRNA prevents binding of aminoglycosides to their target site. At least six distinct genes (rmtA, rmtB, rmtC, rmtD, armA and npmA) encoding their respective methylase enzymes have been reported in P. aeruginosa [52]. MBLs like IMP, VIM, SPM and GIM are frequent in those bacteria. Such enzymes could hydrolyze cephalosporins and carbapenems effectively and their activity was not inhibited by the β-lactamase inhibitors like cavulinic acid and sulbactam but avibactam. Furthermore, mutations in gyrA/B and parC genes mediate resistance to the fluoroquinolones and 98% of Indian superbugs are resistant to nalidixic acid, the 1st DNA gyrase inhibitor used in clinical therapy [53-54].
TB is caused by Mycobacterium tuberculosis and have few related species like M. bovis, M. caprae, M. canettii, M. africanu, M. microti and M. pinnipedii. About 9 million TB cases worldwide and more than half a million women and children have died and about half millions are mdr-TB. Major mdr-tuberculosis burden was found in the three big countries Russia (>46000 cases), China (>61000 cases) and India (>66000 cases). First line oral DOTS Drugs for TB recommended are; 1) Isoniszid; 2) Refampicin; 3) Pyrazinamide; 4) Ethambutol and 5) Streptomycin. For MDR-TB, the recommended drugs are; Kanamycin, Levofloxacin, Cycloserine, Ethionamide, Pyrazinamide, Cycloserine and Ethambutol. For XRD-TB the following drugs in alone or combination has recommended; Capreomycin, PAS, Moxifloxacin, High dose INH, clofazimine, linezolid, and in some cases amoxycillin-clavulanic acid or imipenem-cilastatin type drugs [55-59]. Anti-TB antibiotic refampicin together with isoniazid constitutes the basis of the multidrug treatment regimen for TB. Rifampicin inhibits bacteria RNA synthesis by binding with the β-subunit of the RNA polymerase. The majority of rifampicin-resistant clinical isolates of M. tuberculosis harbour mutations in the 507-533 coding region of rpoB gene creating an altered β-subunit of the RNA polymerase that could not able to bind refampicin. Specifically, codons 516, 526 and 531 are changed to confer refampicin resistance. Isoniazid was introduced in 1952 as an anti-TB agent also known as isonicotinic acid hydrazide. Isoniazid is a pro-drug that requires activation by the catalase/peroxidase enzyme KatG. Isoniazid acts by inhibiting the synthesis of mycolic acids through the NADH-dependent enoyl-acyl carrier protein (ACP)-reductase, encoded by inhA. katG, inhA, ahpC, kasA and NDH rae the genes that may be involved in isoniazid resistance. S315T mutation in katG gene and -15C/T mutation in the promoter may be important in this respect [60].
Streptomycin is a wonderful drug for TB for several decades since its discovery but drug resistant TB now frequent in India. Streptomycin inhibits the initiation of the translation in the 30S ribosome targeting ribosomal protein S12 and the 16S rRNA coded by the genes rpsL and rrs respectively. Consequently, mutations in rpsL (K43R) and rrs (at nucleotide 530 and 915) are the major mechanisms of resistance to streptomycin. Additional mechanisms of resistance are strA gene coding for adenyl transferases that adenylate streptomycin which then could not able to bind ribosome. Fluoroquinolones are currently in use as second-line drugs in the treatment of MDR-TB. Newer-generation quinolones such as moxifloxacin and gatifloxacin are being used as first-line antibiotics with the purpose of shortening the length of treatment in TB. Pyrazinamide was introduced into TB treatment in the early 1950s and constitutes now part of the standard first-line regimen to treat the disease. Pyrazinamide is an analog of nicotinamide and its introduction allowed reducing the length of treatment to six months. Pyrazinamide is also a pro-drug that needs to be converted to its active form, pyrazinoic acid, by the enzyme pyrazinamidase/nicotinamidase coded by the pncA gene. The proposed mechanism of action of pyrazinamide involves conversion of pyrazinamide to pyrazinoic acid, which disrupts the bacterial membrane energetics inhibiting membrane transport. Mutations in the gene pncA gene both in coding region and in the promoter remain as the most common finding in pyrazinamide resistant strains. Second-line anti-TB drugs, linezolid is an oxazolidinone which inhibits an early step in the synthesis of proteins, binding to the 50S ribosomal subunit. Resistance to linezolid in M. tuberculosis is still <2% and mutations in the 23S rRNA and T460C in rplC mutation encoding the 50S ribosomal L3 protein as well as amplification of efflux pumps have been found in many MDR cases [61].
Cholera is caused by oxygenic strains of Vibrio cholerae serogroups O1 and O139. Cholera toxin, encoded by ctxAB in the genome of the lysogenic bacteriophage CTX on the V. cholerae chromosome, is responsible for secretory diarrhea. V. cholerae O139, which emerged in late 1992 in southern India and Bangladesh, is the only non-O1 serogroup that causes epidemic and pandemic cholera. The initial V. cholera O139 isolates, recovered during 1992 to 1993, were sensitive to tetracycline and resistant to trimethoprim-sulfamethoxazole (SXT) and streptomycin. Pan et al. [62] has described an approximately 150 kb conjugative plasmid (pMRV150) as a major contributor to MDR in the O139 strains prevailing in China (accession number: EU116442) which shares a common backbone with pIP1202, an IncA/C conjugative plasmid from a Madagascar MDR isolate of Yersinia pestis and pIP1202-like plasmids found in Salmonella enterica serovar Newport. IncA/C conjugative plasmids in 0139 strains in India that mediates MDR to tetracycline, ampicillin, chloramphenicol, kanamycin, gentamicin, streptomycin, sulfamethoxazole, trimethoprim. Kar et al. [63] in India has shown that the isolated V. cholera El Tor were sensitive to gentamicin, azithromycin and fluoroquinolones; but were resistant to ampicillin, tetracycline, nalidixic acid, furazolidone, streptomycin, erythromycin, co-trimoxazole, neomycin and chloramphenicol suggesting activation of bla, aac, aad and sulI type mdr genes. Folster et al. [64] reported a V. cholera strain 2012EL-2176 harvoring IncA/C2 plasmid containing blaCMY-2, blaCTX-M-2, blaTEM-1, floR, aac(3)-IIa , strA/B, sul1/2, dfrA1, dfrA27, tetA, mphA, mdr-genes and also resistant to ciprofloxacin due to mutation in gyrA(S83I)/parC(S85L). In Africa studies by Quilici et al. [65] and Marin et al. [66] also showed floR, sul2, dfrA1, and strA/B mdr-genes associated with chloramphenicol, streptomycin, sulfamethozaxole and trimethoprim resistance, respectively were found in multi-drug resistant atypical El Tor and Non-O1/Non-O139 V. cholerae [67-69].
Discussion
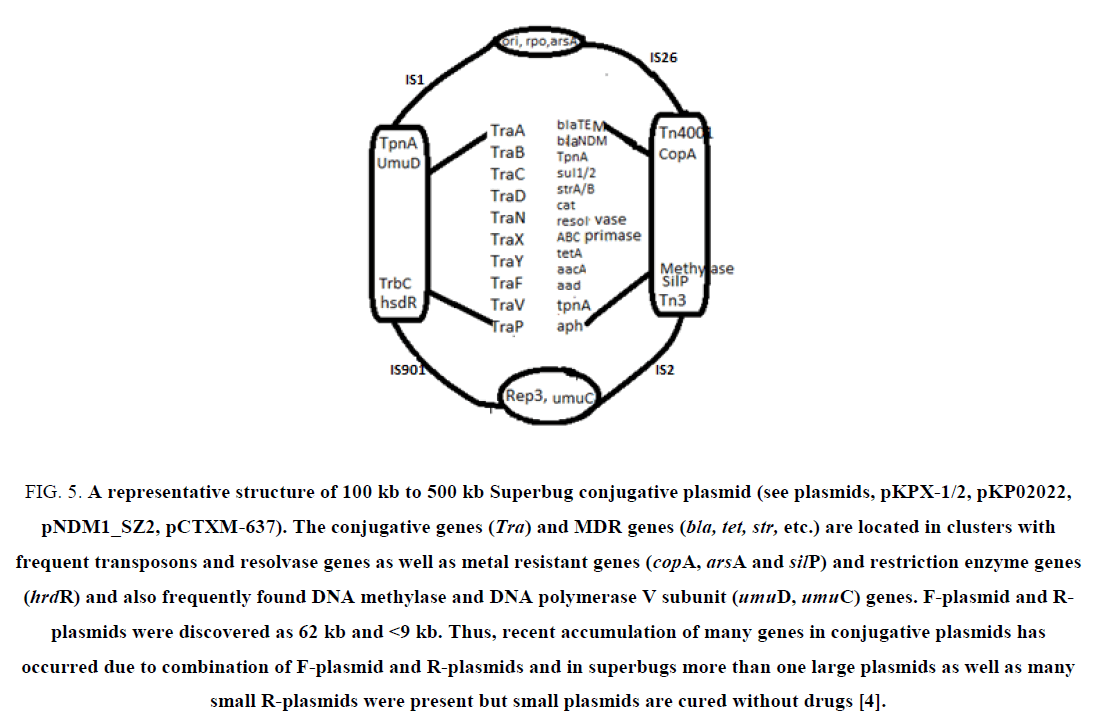
MDR genes are widespread in bacteria and sadly antibiotics are not working to clear infections! Most importantly scientists suggested improper use of antibiotics during therapy (multi-dose 2-3 antibiotics for 3-4 consecutive weeks) as main cause of drug resistance that occurred in the patient’s gastrointestinal tract or blood (Figure 5). Recently, β-lactam inducible bla genes have been discovered and blaI/blaR repressors that control blaZ/blaAmpC genes have been well characterized [70,71]. That mean if you take penicillin which will activates β-lactamases to manifest more MDR and pathogen will survive better, worsening condition of the patient. Further alarming use of antibiotics in agricultural land and in food animal growth has been implicated in the creation of AMR and 2 ng/ml to 20 ng/ml of many antibiotics have been detected in the sea and river water and 100 times excess drug has been reported in Indian and China river near the sewage treatment plant.
Figure 5: A representative structure of 100 kb to 500 kb Superbug conjugative plasmid (see plasmids, pKPX-1/2, pKP02022, pNDM1_SZ2, pCTXM-637). The conjugative genes (Tra) and MDR genes (bla, tet, str, etc.) are located in clusters with frequent transposons and resolvase genes as well as metal resistant genes (copA, arsA and silP) and restriction enzyme genes (hrdR) and also frequently found DNA methylase and DNA polymerase V subunit (umuD, umuC) genes. F-plasmid and R-plasmids were discovered as 62 kb and <9 kb. Thus, recent accumulation of many genes in conjugative plasmids has occurred due to combination of F-plasmid and R-plasmids and in superbugs more than one large plasmids as well as many small R-plasmids were present but small plasmids are cured without drugs [4].
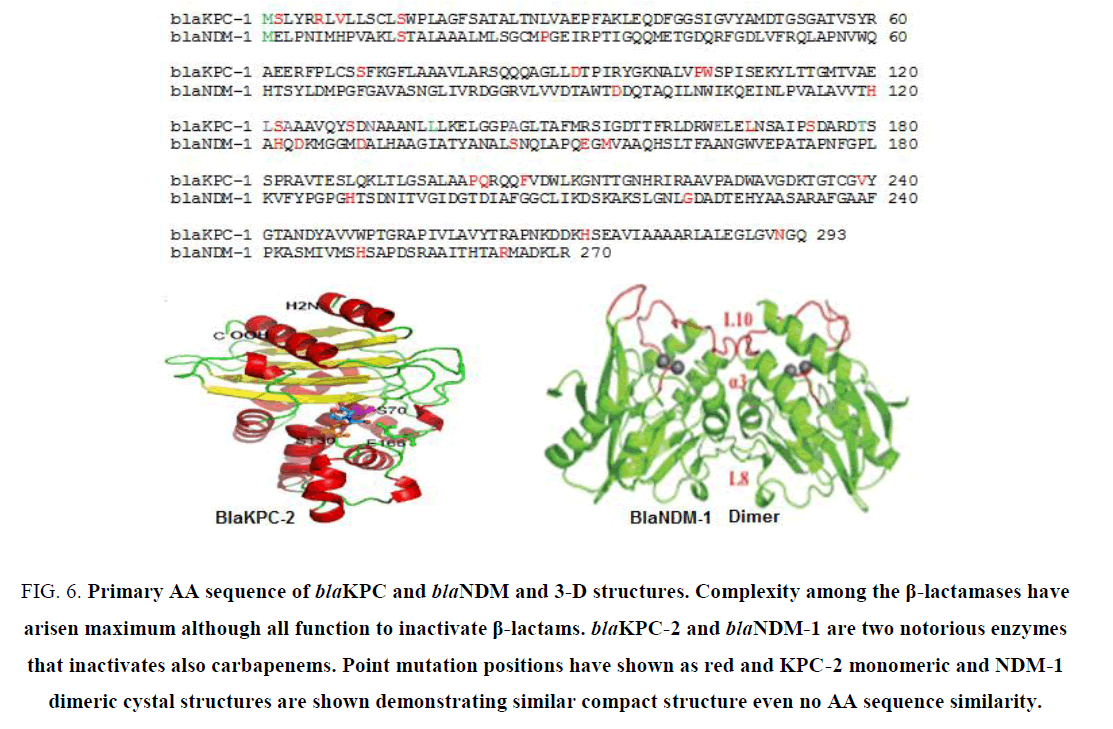
Thousands of MDR genes have been detected and sequenced (Table 1). There are complexity and heterogeneity among the bla genes, aac genes some drug efflux genes [73]. There are hundreds of mutations were found among the same class of MDR gene although such proteins adopted a compact 3-D structures to bind similar substrate. (Figure 6) demonstrated the amino acid sequences of blaKPC-2 and blaNDM-1 and their crystal structure motifs. (Figure 7) demonstrated the primary amino acid sequence of few acetyl-, adenyl- and phospho- transferases showing divergence and heterogeneity to pinpoint vast area of MDR research. The author also excluded drug efflux genes details in this study due to page limit and acrAB, mexAB/CD/EF and tetA/S are very diverged genes and have very potential to exclude many drugs (ampicillin, imipenem, cloxacillin, chloramphenicol, doxacycline, etc. from bacterial cytoplasm to outside including many dye and detergents.
Figure 6: Primary AA sequence of blaKPC and blaNDM and 3-D structures. Complexity among the β-lactamases have arisen maximum although all function to inactivate β-lactams. blaKPC-2 and blaNDM-1 are two notorious enzymes that inactivates also carbapenems. Point mutation positions have shown as red and KPC-2 monomeric and NDM-1 dimeric cystal structures are shown demonstrating similar compact structure even no AA sequence similarity.
Figure 7: Heterogeneity among the drug modifying MDR genes. The primary amino acid sequences of chloramphenicol acetyl transferase, AG acetyl transferase, adenyl transfeases and phospho transferases are presented to demonstrate the diversity of the field of MDR genes which are involved very rapidly due to toxicity of water.
Due to complexity, it is important before therapy to know the drug sensitivities of the pathogen. But so many drug resistant patterns have been observed now-a-day that PCR detection of proper MDR gene isomers has been suggested. Still then, so many PCRs have to be done with hundreds of PCR primers for few hundred MDR genes, that is impractical and expensive. In such scenario, whole genome sequencing of MDR bacteria and large plasmids have been welcomed and merely needs <100 dollars. However, poor peoples in the Indian villages could not afford such expenses of PCR or sequencing or new drugs like colistin, imipenem and investigational drugs like avibactam. India has made MDR monitoring medical board in 2011. Many developed countries like USA and UK are ahead to make National strategy on combating antibiotic resistant bacteria and US alone has granted one billion USD for AMR research in 2016 [4].
Conclusion
Thus, we have concluded that diverse MDR genes have been highly created in household bacteria and now such superbugs are notoriously claiming the lives particularly in immune-compromised neonatal and elderly [74]. As we have to deal with the calamity with new drug discovery but other control measures will be adopted. Therefore, we should aware the release of pollutants (which act as catalyst for MDR gene creation) into environment and also uncontrolled use of antibiotics in human as well as in agricultural farm and in food animal growth. In truth, if the concentration of pollutant (heavy metals, paints and detergents) and antibiotics were reduced, then many drug resistant genes would be vanished from plasmids (due to plasmid rescue mechanism) and in that case, old antibiotics like ampicillin and tetracycline may work again to cure bacterial infection. It is noteworthy to state that without immunological problems, bacteria in our body is controlled by phagocytosis, complement activation and antibody-antigen reactions [75]. It is also true that bacteria help us in the gastrointestinal tract and also in the bio-conversions in the soil and water. Also, bacteria ware evident in air, water and household belongings suggesting synergistic roles in human development. Thus, we must have to reduce populations and industries in big cities to reduce toxic pollutions in air and water. That way we must reduce the new gene creation in bacteria that under stressful condition sometime also creates harmful superbugs like NDM1 E. coli or KPC2 K. pneumoniae [76]. It appears that golden era of antibiotic has ended within 80 years of its discovery. Recently, alternative medicines like heterogeneous herbal antibiotics [77,78], bacteriophage therapy and gene medicines [79-81] are becoming popular all over the world.
Acknowledgement
The author thanks Sourav Nandi, Mitali Maity, Tamanna Roy, Uttam Maity, ex-students of Vidyasagar University for their help during MDR study. The author also thanks Dr. J. B. Medda for financial support.
References
- McArthur AG, Waglechner N, Nizam F. et al. The Comprehensive Antibiotic Resistance Database. Antimicrob Agents Chemther. 2013;57(7):3348-57.
- Lynch JP, Clark NM, Zhanel GG. Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spec¬trum beta-lactamases and carbapenemases). Exp Opini Pharmacother. 2013;14:199-210.
- Okeke IN, Laxminarayan R, Bhutta ZA, et al. Antimicrobial resistance in developing countries. Part I: recent trends and current status. Lancet Infect Dis. 2005;5:481-93.
- Chakraborty AK. High mode contamination of multi-drug resistant bacteria in Kolkata: Mechanism of gene activation and remedy by heterogeneous phyto-antibiotics. Ind J Biotechnol. 2015;14:149-59.
- Ashley EA, Lubell Y, White NJ, et al. Antimicrobial susceptibility of bacterial isolates from community acquired infections in Sub-Saharan Africa and Asian low and middle income countries. Trop Med Int Health. 2011;16:1167-79.
- Drawz SM, Bonomo RA. Three decdes of beta-lactamase inhibitors. Clini Microbiol Review. 2010;23:160-201.
- Bush K, Jacoby GA. Updated functional classification of ß-lactamases. Antimicrob Agents Chemother. 2010;54:969-76.
- Ambler RP. The structure of ß-lactamases. Philosophy Trans Royal Society London. 1980;289:321-31.
- Palzkill T. Metallo-beta-lactamase structure and function. Annals New York Acad Sci. 2013;1277:91-104.
- Jocoby GA. AmpC ß-lactamases. Clini Microbiol Rev. 2009;22:162-82.
- Fosberg KJ, Reyes A, Wang B. et al. The shred antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107-11.
- King DT, Strynadka NC. Targeting metallo-beta-lactamase enzymes in antibiotic resistance. Future Med Chem. 2013;5:1243-63.
- Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. 1940. Rev Infect Dis. 1988;10:677-8.
- Frasson I, Lavezzo E. Antimicrobial treatment and containment measures for an extremely drug-resistant Klebsiella pneumoniae ST101 isolate carrying pKPN101-IT, a novel fully sequenced blaKPC-2 Plasmid. J Clini Microbiol. 2012;50:3768-72.
- Feng Y, Yang P, Wang X, et al. Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J Antimicrob Chemother. 2016;71:71-5.
- Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389-402.
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45-8.
- Shaw KJ, Rather PN, Hare RS, et al. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57(1):138-63.
- Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitation. Biochem Biophys Res Commun. 2014;453:254-67.
- Davis J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417-33.
- Morten O, Sommer A, Dantas G, et al. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325(5944):1128-31.
- Trehan I, Goldbach HS, LaGrone LN, et al. Antibiotics as part of the management of severe acute malnutrition. N Engl J Med. 2013;368:425-35.
- Knox JR. Extended-spectrum and inhibitor resistant TEM-type ß-lactamases: Mutations, specificity and three-dimensional structures. Antimicrob AgentChemother. 1995;39:2593-601.
- Li M, Conklin BC, Tracila MA, et al. Substitutions at position 105 in SHV Family ß-Lactamases decrease catalytic efficiency and cause inhibitor resistance. Antimicrob AgentChemother. 2012;56:5678-86.
- Xu ZQ, Flavin MT, Flavin J. Combating multi-drug resistant gram-negative bacterial infection. Exp OpinInvest Drugs. 2014;23:163-82.
- Wang X, Lu M, Shi Y, et al. Discovery of novel new Delhi metallo-ß-lactamases inhibitors by multistep virtual screening. PLoS One. 2015;10:e0118290.
- Ehmann DE, Jahic H, Ross PL, et al. Kinetics of avibactam inhibition against Class A, C, and D beta-lactamases. J Biol Chem. 2013;288:27960-71.
- McMurry L, Petrucci R, Levy S. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3974-7.
- ChopraI, Robert M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232-60.
- Brenda S, Speer I, Shoemaker NB, et al. Bacterial resistance to tetracycline: mechanisms, transfer and clinical significance . Clini Microbiol Rev. 1992;5(4):387-99.
- Ashenafi M, Ammosova T, Nekhai S, et al. Purification and characterization of aminoglycoside phosphotransferase APH(6)-Id, a streptomycin inactivating enzyme. Mol Cell Biochem. 2014;387:207-16.
- Karmakar S, Biswas D, Shaikh NM, et al. Role of a large plasmid of Salmonella typhi encoding multiple drug resistance. J Med Microbiol. 1991;34:149-51.
- Schwarz S, Kehrenberg C, Doublet B, et al. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev. 2004;28:519-42.
- Chakraborty AK, Hodgson CP. Role of far upstream repressor elements controlling the proto-Ha-ras gene transcription. Biochem Biophys Res Commun. 1998;252:716-22.
- Robicsek A, Strahilevitz J, Jacoby GA, et al. Fluoroquinolone modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med. 2006;12:83-8.
- Guo Q, Weng J, Xu X, et al. A mutational analysis and molecular dynamics simulation of quinolone resistance proteins QnrA1 and QnrC from Proteus mirabilis. BMC Struct Biol. 2010;10:33-6.
- Mataseje LF, Peirano G, Church DL, et al. Colistin non-susceptible Pseudomonas aeruginosa ST654 with blaNDM-1 arrives in North America. Antimicrob AgentChemother. 2016;60(3):1794-800.
- Trobos M, Jakobsen L, Olsen KE, et al. Prevalence of sulphonamide resistance and class 1 integron genes in Escherichia coliisolates obtained from broilers, broiler meat, healthy humans and urinary infections in Denmark. Int J Antimicrob Agents. 2008;32:367-9.
- Johnson TJ, Wannemuehler YM, Johnson SJ, et al. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol. 2007;73:1976-83.
- Wang S, Zhao SY, Xiao SZ, et al. Antimicrobial resistance and molecular epidemiology of Escherichia coli causing bloodstream infections in three hospitals in Shanghai, China. PLoS One. 2016;11(1):e0147740.
- Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-8.
- Reynolds PE. Structure, biochemistry and mechanism of action of glycopeptides antibiotics. Eur J Clini Microbiol Infect Dis. 1989;8:943-50.
- Byarugaba DK. Antimicrobial resistance and its containment in developing countries. In: Gould IM, van der Meer JWM, editors. Antibiotic policies: theory and practice. New York: Springer, USA; 2005. p. 617-47.
- Flanagan JJ, Mukherjee I, Barlowe C, et al. Examination of Sec22 Homodimer Formation and Role in SNARE-dependent Membrane Fusion. J Biol Chem. 2015;290(17):10657-66.
- Merlo TP, Dabul AN, Camargo IL. Different VanA elements in E. faecalis and in E. faecium suggest at least two origins of Tn1546 among VRE in a Brazilian Hospital. Microb Drug Resist. 2015;21(3):320-8.
- Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1 from a carbapenemresistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151-61.
- Papagiannitsis CC, Dolejska M, Izdebski R, et al. Characterisation of IncA/C2 plasmids carrying an In416-like integron with the blaVIM-19 gene from Klebsiella pneumoniae ST383 of Greek origin. Int J Antimicrob Agents. 2016;47(2):158-62.
- Guo Q, Spychala CN, McElheny CL, et al. Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. J Antimicrob Chemother. 2016;71(4):882-6.
- Vila J, Mart S, Sa´nchez-Ce´spedes J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother. 2007;59:1210-5.
- Sun X, Liu B, Chen Y, et al. Molecular characterization of Ambler class A to D ß-lactamases, ISAba1, and integrons reveals multidrug resistant Acinetobacter spp. isolates in north-eastern China. J Chemother. 2016;6:1-7.
- Richmond GE, Evans LP, Anderson MJ, et al. The Acinetobacter baumannii Two-Component System AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio. 2016;7(2):e00430-16.
- Ramakrishnan K, Rajagopalan S, Nair S, et al. Molecular characterization of metallo ß-lactamase producing multidrug resistant Pseudomonas aeruginosa from various clinical samples. Indian J Pathol Microbiol. 2014;57:579-82.
- Sun F, Zhou D, Wang Q, et al. Genetic characterization of a novel blaDIM-2 carrying mega-plasmid p12969-DIM from clinical Pseudomonas putida. J Antimicrob Chemother. 2016;71(4):909-12.
- Yayan J, Ghebremedhin B, Rasche K. Antibiotic resistance of Pseudomonas aeruginosa in pneumonia at a single university hospital center in Germany over a 10-Year period. PLoS One. 2015;10(10):e0139836.
- Halim MZ, Jaafar MM, Teh LK, et al. Genome sequencing and annotation of multidrug resistant Mycobacterium tuberculosis (MDR-TB) PR10 strain. Genom Data. 2016;7:245-6.
- Sloan DJ, Lewis JM. Management of multidrug-resistant TB: novel treatments and their expansion to low resource settings. Trans R Soc Trop Med Hyg. 2016;110(3):163-72.
- Hu Y, Zhao Q, Werngren J, et al. Drug resistance characteristics and cluster analysis of M. tuberculosis in Chinese patients with multiple episodes of anti-tuberculosis treatment. BMC Infect Dis. 2016;16(1):4-8.
- Duan Q, Chen Z, Chen C, et al. The Prevalence of drug-resistant tuberculosis in mainland China: An updated systematic review and meta-analysis. PLoS One. 2016;11(2):e0148041.
- Aye KS, Nakajima C, Yamaguchi T, et al. Genotypic characterization of multi-drug-resistant Mycobacterium tuberculosis isolates in Myanmar. J Infect Chemother. 2016;22(3):174-9.
- He XC, Zhang XX, Zhao JN, et al. Epidemiological trends of drug-resistant tuberculosis in China from 2007 to 2014: A retrospective study. Medicine (Baltimore). 2016;95(15):e3336.
- Garg P, Chakraborty S, Basu I, et al. Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992?1997 in Calcutta, India. Epidemiol Infect. 2000;124:393-9.
- Pan JC, Ye R, Wang HQ, et al. Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob Agents Chemother. 2008;52(11):3829-36.
- Kar SK, Pal BB, Khuntia HK, et al. Emergence and spread of tetracycline resistant Vibr io cholerae O1 El Tor variant during 2010 cholera epidemic in the tribal areas of Odisha, India. Int J Infect Dis. 2015;33:45-9.
- Folster JP, Pecic G, Stroika S, et al. Changing plasmid types responsible for extended spectrum cephalosporin resistance in Escherichia coli O157:H7 in the United States, 1996-2009. J Glob Antimicrob Resist. 2014;2(2):87-91.
- Quilici ML, Massenet D, Gake B, et al. Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, Western Africa. Emerg Infect Dis. 2010;16(11):1804-5.
- Marin MA, Thompson CC, Freitas FS, et al. Cholera outbreaks in Nigeria are associated with multidrug resistant atypical El Tor and non-O1/non-O139 Vibrio cholerae. PLoS Negl Trop Dis. 2013;7(2):e2049.
- Garg P, Sinha S, Chakraborty R, et al. Emergence of fluoroquinolone-resistant strains of Vibrio cholerae O1 biotype El Tor among hospitalized patients with cholera in Calcutta, India. Antimicrob Agents Chemother. 2001;45:1605-6.
- Sameer M, Dixit SM, Johura FT,et al. Cholera outbreaks (2012) in three districts of Nepal reveal clonal transmission of multi-drug resistant Vibrio cholerae O1. BMC Infect Dis. 2014;14:392-6.
- Wang R, Yu D, Zhu L, et al. IncA/C plasmids harboured in serious multidrug-resistant Vibrio cholerae serogroup O139 strains in China. Int J Antimicrob Agents. 2015;45(3):249-54.
- Lindquist S, Lindberg F, Normark S. Binding of the Citrobacter freundii ampR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC ß-Lactamase gene. J Bacteriol. 1989;171:3746-53.
- Llarrull LI, Prorok M, Mobashery S. Binding of the gene repressor BlaI to the bla operon in methicillin-resistant Staphylococcus aureus. Biochemistry. 2010;49:7975-7.
- Kaszubiak A, Holm PS, Lage H. Overcoming the classical multidrug resistance phenotype by adenoviral delivery of anti-MDR1 short hairpin RNAs and ribozymes. Int J Oncol. 2007;31(2):419-30.
- Paulsen IT. Multidrug efflux pumps and resistance: regulation and evolution. Curr OpinMicrobiol. 2003;6:446-51.
- Lubelski J, Konings WN, Driessen AJ. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev. 2007;71:463-76.
- Chakraborty AK. In silico analysis of hotspot mutations in the bacterial NDM-1 and KPC-1 carbapenemases that cause severe MDR phenotypes. Biochem Biotechnol Res. 2016;4(1):17-26.
- Jiang RH, Xu HB, Fu J. Outcomes of Chinese herb medicine for the treatment of multidrug-resistant tuberculosis: a systematic review and meta-analysis. Complement Ther Med. 2015;23(4):544-54.
- Knezevic P, Aleksic V, Simin N, et al. Antimicrobial activity of Eucalyptus camaldulensis essential oils and their interactions with conventional antimicrobial agents against multi-drug resistant Acinetobacter baumannii. J Ethnopharmacol. 2016;178:125-36.
- Lopez C, Arivett BA, Actis LA, et al. Inhibition of AAC (6')-Ib-mediated resistance to amikacin in Acinetobacter baumannii by an antisense peptide-conjugated 2',4'-bridged nucleic acid-NC-DNA hybrid oligomer. Antimicrob Agents Chemother. 2015;59(9):5798-803.
- Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM-1 ß-lactamase. Cell. 2015;160:882-92.
- Chakraborty AK, Zink MA, Boman BM, et al. Synthetic Retrotransposon vectors for Gene Therapy. FASEB J. 1993;7:971-7.
- Chakraborty AK. Complexity. Heterogeneity, 3-D structures and transcriptional activation of multi-drug resistant clinically relevant bacterial beta-lactamases. Trends Biotechnol-Open Access. 2016;2(1):1-001.