Original Article
, Volume: 15( 4)Molecular Docking Studies of the Constituents Present in the Essential Oil of Plectranthus hadiensis against Bacterial Proteins
- *Correspondence:
- Ravi S, Department of Chemistry, Karpagam University, Karpagam Academy of Higher Education Coimbatore, Tamil Nadu, India, Tel: 9047174142; E-mail: ravisubban@rediffmail.com
Received: July 31, 2017; Accepted: August 12, 2017; Published: August 30, 2017
Citation: Sripathi R, Ravi S. Molecular Docking Studies of the Constituents Present in the Essential Oil of Plectranthus hadiensis against Bacterial Proteins. Int J Chem Sci. 2017;15(4):185
Abstract
Plectranthus is a large and widespread genus with a diversity of the ethnobotanical uses. In traditional medicine, the juice of stem and leaves of Plectranthus hadiensis when mixed with honey, is used as a remedy for diarrhea. The chemical composition of the essential oil of Plectranthus hadiensis is determined by GC-MS analysis and the antimicrobial efficacy of the oil has evaluated in the previous studies. The aim of the present study is to carryout molecular docking of the essential oil constituents against the bacterial proteins present in murD ligases 1UAG, 2X5O, 3UDI and 3TYE a dihydropteroate synthase enzyme. Caryophyllene, germacrene and cubebene are found to be more effective. These compounds act on multi targets and serve as antibacterial agents.
Keywords
Plectranthus; Molecular docking; Protein; Caryophyllene
Introduction
Essential oils are characterized by a strong odour and are a mixture of natural, complex volatile compounds. Essential oils are used invariably in the preservation of foods, sedative, spasmolytic, analgesic, anti-inflammatory and local anesthetic remedies [1]. They have a special fragrance and known for their microbial properties [2]. So, they find applications as naturally occurring antimicrobial agents in pharmaceutical botany, pharmacology, phytopthalogy, food preservation, medical and clinical microbiology.
Plectranthus genus consists of 300 species, distributed from Africa to Asia and Australia [3,4]. This plant belongs to the Lamiaceae family and is used in Ayurveda. In India, it is found in all the habitats, especially in the Himalaya, the Southern Western Ghats, and the Nilgiris region. The studies on the chemical constituents and biological activities of Plectranthus species growing in India are scarce [5]. Available reports on this genus revealed that Plectranthus species present in India are richer in essential oil content. The essential oil composition has been reviewed and reported [6,7]. The pharmacognostic characteristics, phytochemical analysis and the compounds 7β-Acetoxy-6β-hydroxyroyleanone, 7β-Acetoxy-6β-hydroxyroyleanone and 6β, 7β-dihydroxyroyleanone are reported from P. hadiensis [8]. Only recently, the essential oil composition has been reported by us. Since high-throughput screenings have continued to show disappointing results, the present practice of drug development has slowly shifted towards the structure-based drug design. By investigating the ligand interactions with its receptor proteins, the space between hit generation and drug establishment can be narrowed down. Therefore, the knowledge of molecular level interactions between specific protein target and developed ligands will be highly beneficial in the drug development research, since this information can be used to design new molecules with improved protein fitting. The objective of the present study is to carry out molecular docking of the essential oil constituents against the bacterial proteins [9-20].
Material and Methods
Target proteins
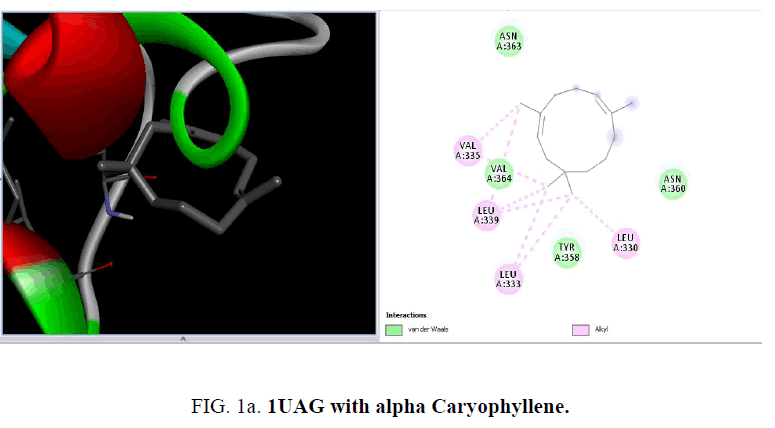
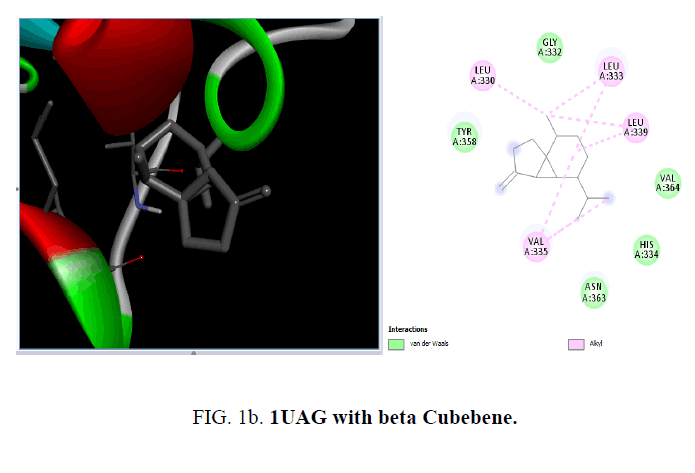
From the RCSB Protein Data Bank the three-dimensional structure of the proteins 1UAG, 2X5O and 3UDI and 3TYE were obtained for the present study (FIG. 1a and FIG. 1b).
Ligand selection
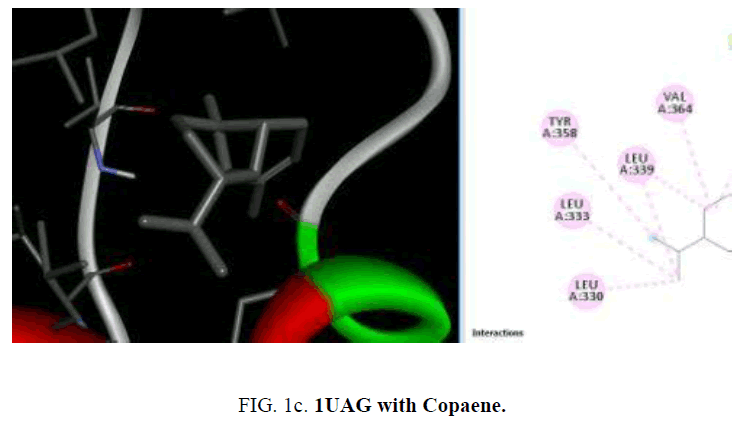
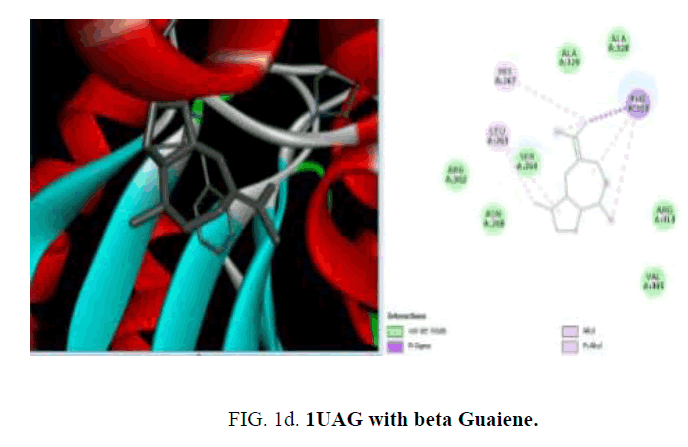
The compounds present in the essential oil obtained from P. hadiensis was selected for docking studies. The structure of the compounds was drawn using Chem draw and MOL SDF format and to generate atomic coordinates they were converted to PDBQT file using PyRx tool (FIG. 1cand FIG. 1d).
Optimization of the target and the ligand
The PDB coordinates of the target protein and ligand molecules were optimized using UCSF Chimera tool and Drug Discovery Studio version 3.0 software because these coordinates had a minimum energy and stable confirmation.
Target active binding site analysis
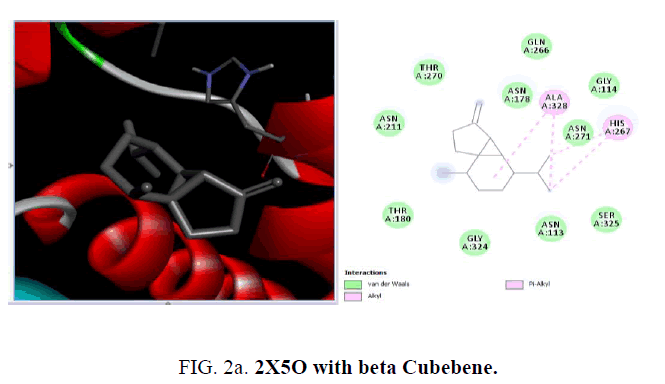
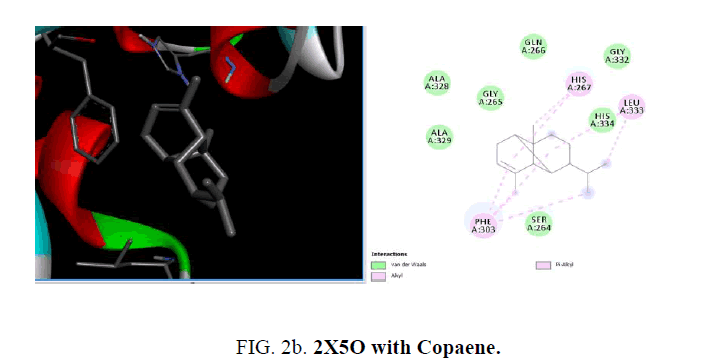
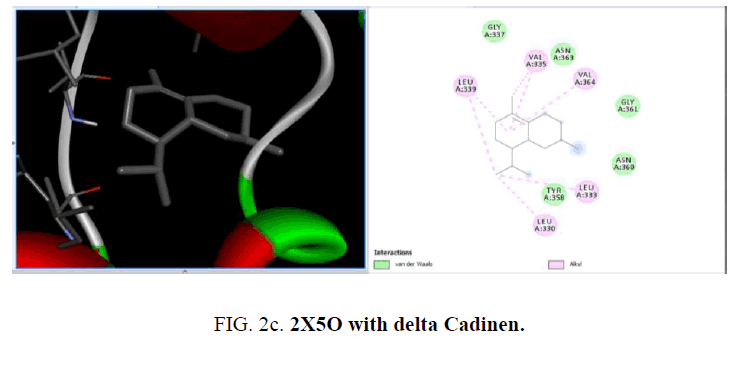
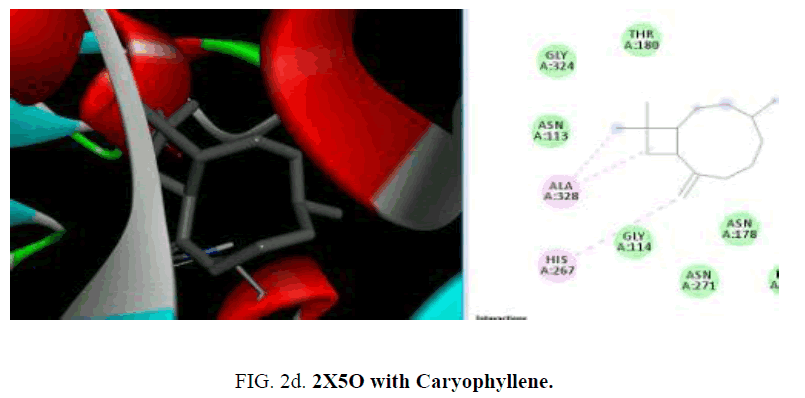
The active sites were the coordinates of the ligand in the original target protein grids, and these active binding sites of target protein were analyzed using the Drug Discovery Studio version 3.0 and 3D Ligand Site virtual tools (FIG. 2a -2d ).
Molecular docking analysis
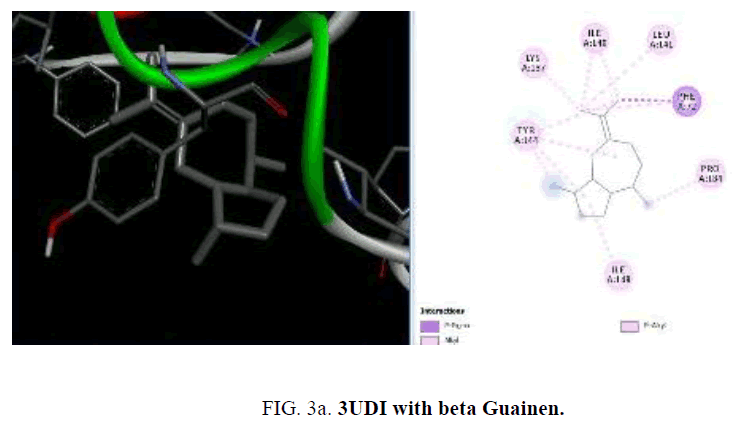
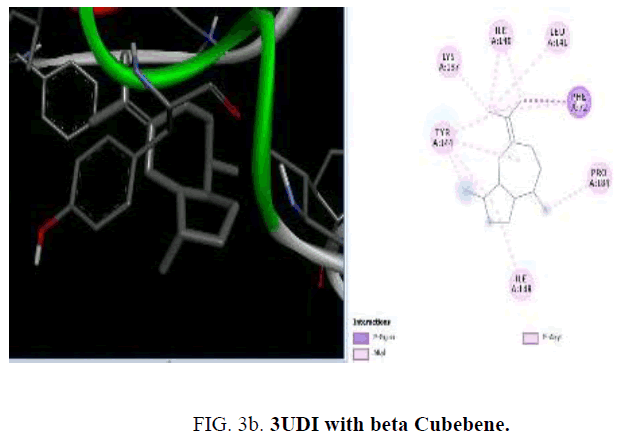
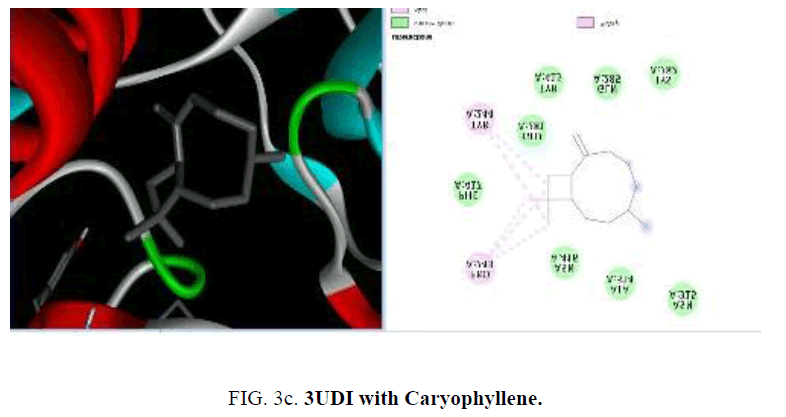
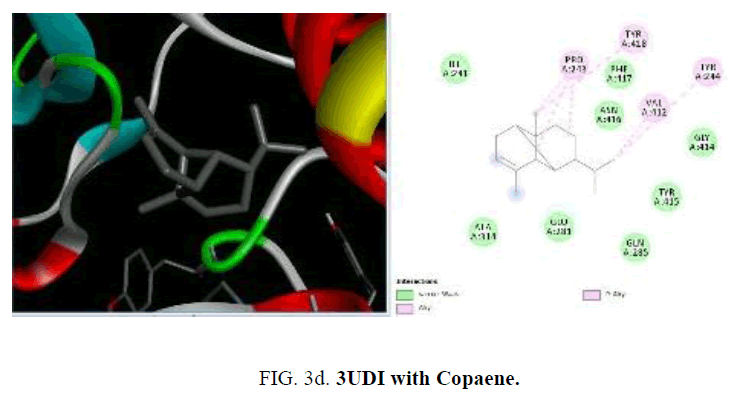
To start with protein-ligand interaction was analyzed for hydrophobic and hydrophilic properties of these complexes using Platinum software web server. Then a computational ligand-target docking approach was used to study the structural complexes of the proteins 1UAG, 2X5O and 3UDI and 3TYE (target) with the compounds present in the essential oil (ligand) in order to study the structural basis of these protein target specificity. Finally, docking was carried out by PyRx, AutoDock option based on scoring functions (FIG. 3a-3d).
Results and Discussion
Many organisms showed drug resistance due to inadequate use of antibiotics and cause damage to human health. So, a need arises for finding new substances from natural sources, including plants. Antibacterial activity of the essential oils of P. hadiensis was tested against Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa by using zone of inhibition method and appeared to be very active. The major constituents of the essential oil obtained from the seed and aerial parts are piperitone oxide, copaene, β-farnesene, α-caryophyllene, 3, 5-heptaddienal, 2-ethylidene, 6-methyl, p-cymen-3-ol, L-fenchone and limonene.
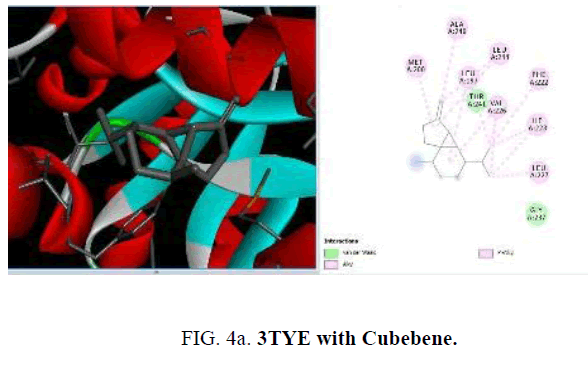
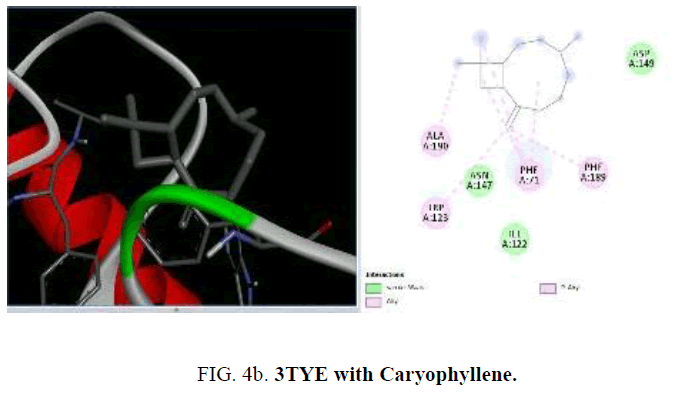
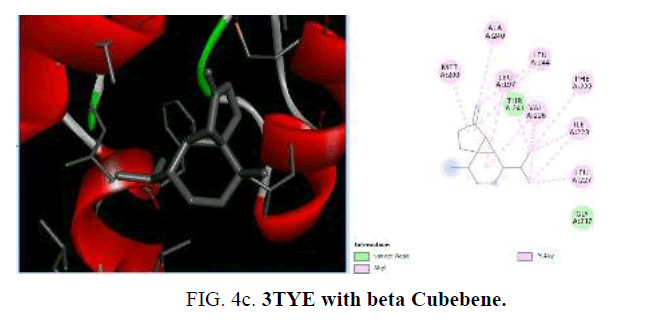
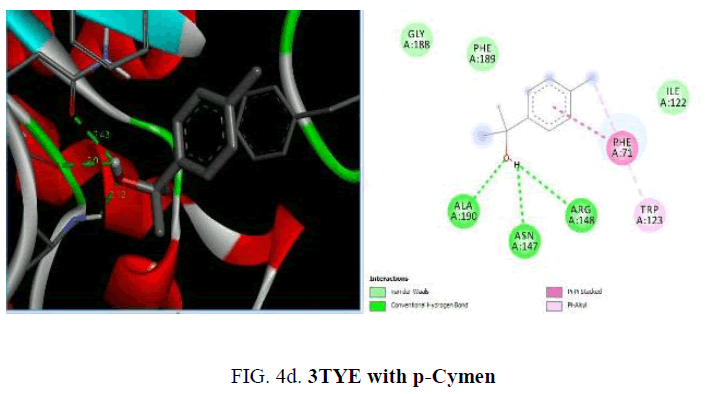
Now, in the current study, the knowledge on the target proteins of currently used antibiotics is extended to the phytoconstituents which is identified from P. hadiensis. Docking studies are performed for all the identified compounds from the essential oil of P. hadiensis in order to examine their affinity with the bacterial proteins that are well known targets for some antibiotics with different mechanism of action such as cell wall synthesis, inhibitors of nucleic acid synthesis and antimetabolites (FIG. 4a-4d).
The proteins used are present in the enzyme murD ligase (1UAG, 2X5O and 3UDI) which involve in the cell wall synthesis and dihydropteroate synthase enzyme (DHPS; PDB id 3TYE). The essential oil constituents are docked to the active sites of each protein using AutoDock4 in order to validate the docking approach for the protein structures used. Some protein structures presented either natural substrates or known inhibitors as co-crystallized ligands. Each co-crystallized ligand is removed from the respective protein before docking and scoring validation process is used. The docking score, hydrogen bonded residues, and hydrophobic interactions like alkyl and pi alkyl, van der Waals interactions are provided. Most of the compounds showed very good interactions with the studied proteins.
Mur ligases (Mur C, D, E and F) are used in the synthesis of the peptide stem of bacterial peptidoglycan. Among the four enzymes MurD is responsible for the ATP-dependent addition of d-glutamic acid to UDPMurNAc-1-ala, a reaction which involves an acyl-phosphate and tetrahedral intermediates. Thus, MurD inhibitors would tend to disturb the cell wall synthesis of microorganisms, and exhibit antimicrobial potential. The crystal structure of MurD ligase (PDB ID: 1UAG) indicated the presence of three globular domains-D1, D2 and D3.
D1 is composed of a five-stranded parallel β-sheet, surrounded by four α-helices and primarily responsible for the fixation of UDP moiety of UMA. D2 is formed of a central six stranded β-sheet, surrounded by seven α-helices, and a small flanking three stranded antiparallel β-sheet. D3 is composed of a six stranded β-sheet with parallel and antiparallel strands, and five neighboring α-helices. On the cellular level, the substrate molecule (UMA) binds to MurD ligase in the cleft architecture between D1 and D2. Like most enzymes of peptidoglycan biosynthesis, MurD constitutes an attractive target for the design and synthesis of new agents. Among the essential oil constituent’s studies (TABLE 1), with the protein 1UAG caryophyllene (-6.1 Kcal/mol), germacrene (-6.8 Kcal/mol), guaiene (-6.3 Kcal/mol), cubebene (-6.4 Kcal/mol) and copane (-6.4 Kcal/mol) show good docking scores when compared to other compounds due to hydrophilic and hydrophobic interactions. None of them show hydrogen bonded interactions with the protein. Mostly all the above compounds show interactions with LEU: 339, LEU: 333 and LEU:330.
| Ligand | Scouring | H-bond | Alkyl and pi-alkyl | Others |
|---|---|---|---|---|
| Alpha-Caryophyllene | -6.3 | - | VAL:335, LEU: 339,333,330 | - |
| beta-Guaiene | -6.3 | - | LEU: 263, HIS: 267, PHE:303 | Pi-sigma PHE:303 |
| beta-Cubebene | -6.4 | - | LEU:3330,333,339, VAL:335 | - |
| Cis-p-Methyl-2, 8-Diene-1-ol | -5.4 | TYR:358 | VAL: 335,364, LEU: 339,333 | - |
| Copaene | -6.4 | - | VAL: 364,335, TYR:358, LEU:339, 333,330 | - |
| D-Limonene | -5.0 | - | LEU:333, PHE:303, HIS:267 | - |
| Delta-Cadinene | -6.3 | - | ARG:302, PHE:303, HIS:267, LEU:263 | - |
| Diosphenol | -5.9 | ALA:328, ASN:268 | LEU:333, PHE:303 | (C-H bond) ALA:329 |
| Beta-Farnesene | -4.7 | - | VAL:364, TYR:358, LEU: 339,333,330 |
- |
| L-Fenchone | -5.4 | 1 | LEU: 339,333,330, VAL: 364,335 | - |
| Naphthalene, 1, 2, 3, 4, 4a,7-hexahydro-1, 6-dimethyl-4-(1-methylethyl | -6.2 | - | PHE:303, HIS:267, LEU:333 | - |
| p-Cymen-3-ol | -5.7 | ASN:268 | LEU:333, PHE:303 | Pi-pi stacked, PHE:303, unfavourable acceptor, SER:264 |
| Pipertone oxide | -5.2 | ASN:268, HIS:267 | PHE:303, LEU:333 | |
| Thymol | -5.2 | ASN:268 | ALA:329, PHE:303, HIS:267 | Pi-pistcked, PHE:303, pi-cation, HIS:267 |
| trans-m-Propenyl guaiacol | -5.5 | SER:264, ASN:268 | LEU:333 | Pi-cation, HIS:267, pi-pistcked,PHE:303 |
| Pulespenone | -5.6 | HIS:267, ASN:268 | LEU:333, HIS:267, PHE:303 | Pi-sigma, PHE:303 |
| Germacrene D | -6.1 | - | TYR:358, VAL:364, LEU:333, LEU:330, LEU:339 | GLY:365, ASN:360 |
TABLE 1. Docking study of the essential oil constituents from P. hadiensis against 1UAG protein.
The crystal structure of MurD ligase (PDB ID: 2X5O) also reveals the presence of three globular domains-D1, D2, and D3 and it is structurally similar to 1UAG. Among the phytoconstituents studies (TABLE 2), with the protein 2X5O caryophyllene (-6.6 Kcal/mol), germacrene (-6.6 Kcal/mol), cubebene (-6.4 Kcal/mol), δ-cadinene (-6.3 Kcal/mol) with and copane (-6.1 Kcal/mol) show good docking scores when compared to other compounds due to hydrophilic and hydrophobic interactions. Consistent with the result from the protein 1UAG here also none of them show hydrogen bonded interactions with the protein and most of the compounds show interactions with LEU:339, LEU:333 and LEU:330. The absence of H-bonded interactions is due to the scarcity of polar groups in the ligands.
| Ligands | Scouring | H-bond | Alkyl and pi-alkyl | Others |
|---|---|---|---|---|
| Alpha-Caryophyllene | -6.1 | - | ALA:328, HIS:267 | - |
| Beta Cubebene | -6.1 | - | ALA:328, HIS:267 | - |
| Bisabolol | - | ALA:328, ASN:331 | HIS:267, PHE:303 | - |
| Camphor | -5.0 | - | LEU:416, PHE:422, ALA:414 | - |
| Copaene | -6.1 | - | HIS:267, LEU:333, PHE:303 | - |
| D-Limonene | -5.1 | - | LEU: 339,330,333, TYR:358, VAL:335 | - |
| Delta-Cadinene | -6.3 | - | LEU: 330,333,339, VAL: 335,364 | - |
| L-Fenchone | -5.1 | VAL:335 | LEU: 339,333, VAL:364 | - |
| Naphthalene, 1, 2, 3, 4, 4a, 7-hexahydro-1, 6-dimethyl-4-(1-methylethyl) | -6.3 | - | PHE:303, HIS:267, ALA:329, LEU:333 | - |
| Germacrene D | -6.6 | - | LEU:333, 330, 339, VAL: 364, TYR:350 | VAL:335, ASN:363 |
TABLE 2. Docking study of the essential oil constituents from P. hadiensis against 2X5O protein.
Protein binding proteins (PBPs) are biosynthetic enzymes of bacterial cell wall assembly that are found anchored in the cell membrane and are involved in the crosslinking of bacterial cell wall. β-lactam antibiotics target the penicillin binding proteins. They are responsible for transpeptidation, transglucosylation and carboxypeptidation reactions. They are involved in the assembly, maintenance and regulation of peptidoglycan structure in both Gram-positive and Gram-negative bacteria. All these proteins are involved in the final stages of the synthesis of peptidoglycan, which is the major component of bacterial cell walls. Bacterial cell wall synthesis is essential to growth, cell division (thus reproduction) and maintaining the cellular structure in bacteria. Inhibition of PBPs leads to an irregularity in cell wall structure such as elongation, lesions, loss of selective permeability, and eventual cell death and lysis. One such protein is with PDB id: 3UDI is used in the present study (TABLES 3 and 4).
| Ligand | Scouring | H-bond | Alkyl and pi-alkyl | Others |
|---|---|---|---|---|
| 2, 3-Dimethylhydro quinone | -6.1 | VAL:649 | ARGl:482, TYR:485 | (C-H Bond) VAL;649 PiAnionASP:648 Pi-pi stacked TYR:485 |
| α-Caryophyllene | -6.6 | - | ALA:66, PRO:184, PHE:63,72, LEU:141, TYR:144 | - |
| β-Guaiene | -7.1 | - | LYS:137, ILE;140, LEU:141, 148, TYR:144, PRO:148 |
Pi-sigma PHE:72 |
| β-Cubebene | -6.8 | - | LEU:141, ILE:148, 140, PHE:63,72 PRO:184, TYR:144 |
- |
| Bisabolol | -6.3 | - | ILE:645, LEU:486, TYR:485ARG481 482, VAL;649 |
- |
| Copaene | -7.0 | - | PRO:243, VAL:412, TYR: 244,418 | - |
| D-Limonene | -6.2 | - | VAL:649 ILE:645 ARG: 481,482 | TYR:485(pi-sigma) |
| 2 ,3, 4, 4a, 5, 6, 6a, 7, 8, 9-Decahydropyrano [3, 2-H] 3Chromen-5 , 5, 6, 6-tetracarbonitrile |
-7.5 | GLY: 709,708 THR:672 SER:487 |
TYR; 485,707 | - |
| Delta-Cadinene | -6.6 | - | LYS:137 ILE: 140,148, PHE:72 LEU:141 PRO:184 |
TYR:144 |
| Diosphenol | -6.3 | GL:281 GLN:285 | TYR:418, 244 PRO:243 VAL:412 |
- |
| α-Elemene | -6.2 | - | TYR:144, ILE:140148, PHE:72LEU:141 PRO:184 |
- |
| Endo-Fencyl alcohol | -6.3 | - | ARG:481, VAL:649, TYR:485 HIS:652 |
Arg:482 (unfavourable donor) |
| Beta-Farnesene | -5.9 | - | TYR:485, ARG:482, LEU:486 VAL:649 ILE:645 |
- |
| 6-(3-Isopropenylcycloprop-1-enyl) | -6.0 | LYS:137 | ILE:140, TYR:144 | PHE:72(pi-sigma) |
| L-Fenchone | -5.5 | - | PHE:72, LYS:137, ILE:140, LEU:141 | TYR:144(pi-SIGMA) |
| Naphthalene, 1, 2, 3, 4, 4a, 7-hexahydro-1, 6-dimethyl-4-(1-methylethyl) | -7.4 | - | ILE140, LYS;137 PHE: 72,148 PRO:184 |
TYR:144(Pi-SIGMA) |
| p-Cymen-3-ol | -6.0 | - | ILE:645, VAL:649ARG:487 | Pi-anion ASP:648 Pi-Pi TYR:485 |
| Piperitone oxide | -5.6 | ASN:416 | PRO:243, TYR:244 VAL:412 | ALA:314(c-HBOND) |
| Terpinolene | -6.5 | - | TYR:485, ILE:645, VAL:649, ARG:487 | - |
| Thymol | -6.3 | - | ARG: 482,481, VAL:649, ILE:645 | Pi-ANION ASP:648 Pi-Pi STAKED TYR:485 |
| Pulespenone | -6.7 | - | VAL:649, TYR:485ARG481ILE:645 | - |
TABLE 3. Docking study of the essential oil constituents from P. hadiensis against 3UDI protein.
| Ligand | Scouring | Alkyl and pi-alkyl |
|---|---|---|
| Cubebene | -6.6 | MET:200, ALA:240, LEU: 197,244,227, VAL:226, ILE:223, PHE:22 |
| D-Limonene | -4.8 | PHEl: 71,191, TRP:123 |
| Delta-Cadinene | -5.3 | ILE:272, LYS:274,2 |
| Germacrene D | -6.4 | ILE:223, LEU:227, 244, 197, VAL:226 |
TABLE 4. Docking study of the essential oil constituents from P. hadiensis against 3TYE protein.
Among the phytoconstituents studies, with the protein 3UDI caryophyllene (-6.6 Kcal/mol), guainen (-7.1 Kcal/mol), germacrene (-6.6 Kcal/mol), cubebene (-6.4 Kcal/mol), δ-cadinene (-6.6 Kcal/mol), elemene (-6.2 K Cal/mol), fencyl alcohol (-6.3 Kcal/mol), pulespenone (-6.7 Kcal/mol), thymol (-6.3 Kcal/mol), terpenolene (-6.5 Kcal/mol), bisabolol (-6.3 Kcal/mol), and copane (-7.0 Kcal/mol) show good docking scores when compared to other compounds due to hydrophilic and hydrophobic interactions germacrene shows hydrophobic interactions with TYR:244, LYS:222, VAL:412 and LEU:339. Cubebene indicates the hydrophobic interactions with ELU:141, ILE:148,140, TYR:144, PRO:184 PHE:72,63, thymol exhibits hydrophobic interactions with ARG:482,481, VAL:649, ILE:645, pulespenone exhibites hydrophobic interactions with VAL:649, TYR:485 ARG481 ILE:645 and copane shows hydrophobic interactions with PRO:243, VAL:412 and TYR:244,418. None of them show hydrogen bonded interactions with the protein. The site of interaction between the compounds and the protein differs from one compound to another compound.
The sulfonamide antibiotics inhibit dihydropteroate synthase (DHPS), a key enzyme in the folate pathway of bacteria and primitive eukaryotes. It is known that sulfonamides reduce the biosynthesis of dihydrofolic acid through the competitive inhibition of the dihydropteroate synthase enzyme (DHPS). The crystal structure of dihydropteroate synthase enzyme (DHPS; PDB id 3TYE) is also used in the present study. Among the phytoconstituents studies, with the protein 3TYE the compounds caryophyllene (-6.2 Kcal/mol), germacrene (-6.4 Kcal/mol), cubebene (-6.6 Kcal/mol), guaine (-6.1 Kcal/mol) and P-cymen-3-ol (-6.6 Kcal/mol) show good docking scores when compared to other compounds due to hydrophilic and hydrophobic interactions. Caryophyllene forms hydrophobic interactions with ALA: 190, PHE: 71, 189, TRP: 123. Germacrene shows hydrophobic interactions with ILE: 2 23, LEU: 227, 244, 197, VAL: 226 and cubebene indicates the hydrophobic interactions with MET: 200, ALA: 240, LEU: 197, 244, 227, VAL: 226, ILE: 223, PHE: 22. None of them form H-bonds with the protein. Among the essential oil constituent’s studies, against the proteins, (1UAG, 2X5O and 3UDI) dihydropteroate synthase enzyme (DHPS, 3TYE), the compounds caryophyllene, germacrene and cubebene are found to be more effective. These compounds act on the multi targets and may serve as antibacterial agents. They involve in different types of mechanisms and act as, inhibitors of nucleic acids synthesis and as MurD ligase inhibitors which involve in cell well synthesis.
Conclusion
Molecular docking studies was carried out for the essential oils constituents against the bacterial proteins (1UAG, 2X5O and 3UDI) and (3TYE). Caryophyllene, germacrene and cubebene were found to be more effective. These compounds act on multi targets and serve as an antibacterial agent.
References
- Burt S. Essential oils: Their antibacterial properties and potential applications in foods a review. J Food Microbiol. 2004;94:223-53.
- Hammer KA, Carso CF, Riley TV. Antimicrobial activity of essential oils and other plant extracts. J App Microbiology. 1999;86:985-90.
- Codd LE. In: Flora of Southern Africa. Balogh Scientific Books. 1985;28:37.
- Weyerstahl P, Kaul VK, Meier N, et al. Volatile Constituents of Plectranthus rugosus leaf oil. Planta Med. 1983;48:99.
- Ammon HP, Kemper FH. Ayurveda: 3000 years of Indian traditional medicine. Med Welt. 1982;33:148-53.
- De Souza NJ, Shah V. An adenylate cyclase activalmgurug from an Indian herb. Plant Res. 198;2:1.
- Pokhria S, Joshi BC, Pathak U. Genus Plectranthus in India and its chemistry. Chem Bio Div. 2011;8(2):244-52.
- Mehrotra RA, Vishwakarma RS. Abietane diterpenoids from Coleus Zeylanicus. Phytochem. 1989;28:3135-7.
- Brugueras M, Carcia M, Diaz R. Actualidad de las quinolonas. Rev Cubana Farm. 2005;39:1-15.
- Murry PR, Rosenthal KS, Pfaller MA. Microbiologia Medica. 5th ed. Mosby, Elsevier, Rio De Janerio, Brazil. 2006.
- Joshi RK. Chemical composition and antimicrobial activity of the essential oil of Plectranthus mollis (Lamiaceae) from Western Ghats Region, Karnataka, India. Chem Bio Div. 2011;8(2):244-52.
- Pal M, Kumar A, Tewari SK. Chemical composition and mosquito repellent activity of the essential oil of Plectranthus incanus, Facta universitatis series: Physics. Chem Tech. 2011;9(1):57-64.
- Karousou R, Lanaras T, Kokkini S. Piperitone oxide-rich essential oils from Mentha longifolia sub sp. petiolata and M. x villoso-nervata grown wild on the Island of Crete (S Greece). J Esst Oil Res. 1998;10:375-3.
- Turkez H, Togar B, Tatar A, et al. Cytotoxic and cytogenetic effects of α-copaene on rat neuron and N2a neuroblastoma cell lines. Biologia. 2014;69:936-42.
- Radulovic NS, Blagojevic PD, Stojanovic-Radic ZZ. Antimicrobial plant metabolites: Structural diversity and mechanism of action. Curr Med Chem. 2013;20:932-52.
- Nazzaro F, Fratianni F, Martino LD, et al. Effect of essential oils on pathogenic bacteria. Pharmaceuticals (Basel). 2013;6(12):1451-74.
- Huang L, Crothers K, Atzori C, et al. Dihydropteroate synthase gene mutation in pnemocystis and sulfa resistance. Energy Infect Dis. 2004;10:1721-8.
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: From targets to networks. Nat Rev Microbiol. 2010;8:423-35.
- Faleiro ML. The mode of antibacterial action of essential oils, science against microbial pathogens: Communicating current research and technological advances. A Mendez-Vilas. ed. 2011;pp:1143-56.
- Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006;119:S3-S10.