Original Article
, Volume: 14( 5)Identification of the Suitable Model Organism for Human Caspases Study Using Bioinformatics Approach
- *Correspondence:
- Jinny Tomar , Amity Institute of Biotechnology, Amity University, Haryana, India, E-Mail: jinny.tomar@gmail.com
Received: June 13, 2017; Accepted: July 06, 2017; Published: October 05, 2018
Citation: Tomar J, Sharma N. Identification of the Suitable Model Organism for Human Caspases Study Using Bioinformatics Approach. Biotechnol Ind J. 2018;14(5):172.
Abstract
Caspases are critical for a cell functioning. To understand the functioning of human caspases different model organisms could be considered. Our study suggests that human executioner caspases, initiator caspases and inflammatory caspases shows different level of association with the model organisms under study. This study suggests that for human caspases related studies either Pan troglodytes or Felis catus should be considered based on the extent of sequence similarity. This clearly indicates that we should rethink while performing experiments on Mus musculus, as it may not show the same level of association, thereby may lead to decrease in the efficacy of drug designed for humans and tested on mouse.
Keywords
Model organism; Human caspases; Bioinformatics approach
Abbreviations
Casp: Caspase; PCD: Programmed Cell Death; DED: Death Effector Domain; CARD: Caspase Recruitment Domain; FADD Fas Associated Death Domain; ICE: IL-1 beta Converting Enzyme; APAF: Apoptosis Protease Activating Factor; NEDD: NEURAL Precursor Cell Expressed Developmentally; MCH: Melanin-Concentrating Hormone Receptor; CAP: Catabolite Activator Protein; FLICE: FADD-like IL-1beta-Converting Enzyme; ICE: LAP6(ICE-Like Apoptotic Protease6); PPP1R569: Protein Phosphatase 1 Regulatory Subunit 56; ALPS2: Autoimmune Lymphoproliferative syndromes2; NCBI: National Center for Biotechnology Information; T-coffee: Tree-based Consistency Objective Function For alignment Evaluation; MUSCLE - Multiple Sequence Comparison by Log-Expectation; POWER- Power Phylogenetic Web Repeater
Introduction
Caspases are intracellular proteases that cause apoptosis, inflammation, necrosis [1]. Caspases are cysteine proteases that cleave the proteins at specific aspartate residues [2].
Apoptosis
Apoptosis or Programmed Cell Death (PCD) is a naturally occurring process in the body in which cells are damaged by an injurious agent. Apoptosis is important for normal development and tissue homeostasis, but too much or too little apoptosis can also cause disease. The family of cysteine proteases (caspases) is critical mediators of programmed cell death. Apoptosis is important in all stages of life. For example, during embryonic development apoptosis ensures that undesirable or outmoded tissues disappear. Those biochemical events which cause changes in cell and death are blebbing, cell shrinkage, and nuclear fragmentation.
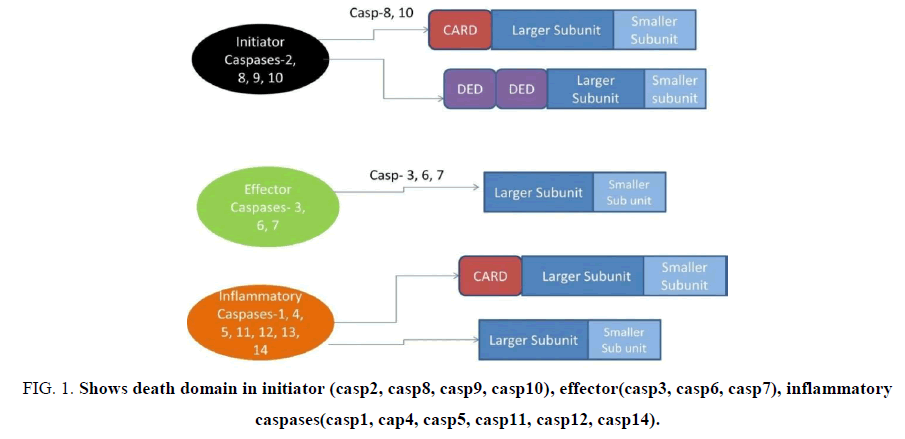
Type of Caspases: Caspases are divided into three groups (Fig. 1)
Figure 1: Shows death domain in initiator (casp2, casp8, casp9, casp10), effector(casp3, casp6, casp7), inflammatory caspases(casp1, cap4, casp5, casp11, casp12, casp14).
1. Initiator caspase
2. Effector caspase
3. Inflammatory caspase
These caspases consist of death domains as shown below in Fig. 1[3]. It consists a death domain.
Initiator Caspases (Casp2, Casp8, Casp9, Casp10): Caspases which have long prodomain are believed to be upstream initiator caspases. Casp 8 and 10 contain two DED domains within their prodomain. Homotypic interactions occur between DEDs of Casp 8 and 10 and DEDs of adapter molecule, FAS- Associating Protein with Death Domain (FADD), induce the recruitment of caspases to death receptors and lead to caspase activation. Casp 1,4,9,2 consists of CARD domain. CARDs of these caspases interact with CARD-containing adapter molecules. These caspases activate the effector caspases.
Initiator apoptotic caspases and inflammatory caspases contain prodomains of over 100 amino acids, while the prodomains in effector caspases are usually less than 30 amino acids. The short prodomains of executioner caspases are unlikely to mediate protein-protein interactions. Rather, they seem to inhibit caspase activation [4]. Initiator caspases: casp2, casp8, casp9, casp10. These caspases activate the effector caspases.
The release of cytochrome c from mitochondria due to cellular stress and presence of dATP, stimulates Apoptosis protease activating factor 1 (Apaf1) which leads to the formation of the apoptosome. This heptameric complex of Apaf1, CARD, and ATPase domain further activates the inactive caspase 9, which thereby initiates apoptosis [5].
Effector caspases: Caspases 3, 6, 7 contain short prodomain and are known as downstream caspases which are activated by upstream caspases. When the C-terminal, smaller subunit of the protease domain is released, then, the prodomain is removed from the large subunit of the protease. Crystallography studies suggest that active caspases are heterotetramers and are composed of two large subunits and two small subunits [6]. Activation of executioner caspase is done by initiator caspases. The cleavage between the large and small subunits allows a conformational change that brings the two active sites of the executioner caspase dimer together and creates a functional mature protease [7].
Inflammatory caspases (casp4, casp5, casp1, casp12): The human inflammatory caspases include caspase-1 (IL-1 beta converting enzyme (ICE), -4, - 5, and -12. The role of caspase-1 in the maturation of IL-1beta and IL-18 it plays a key role in response to pathogenic infection as well as in inflammatory and autoimmune disorders. In case of caspases 1, the activating inflammasome is commonly known as Nucleotide Binding And Oligomerization Domain (NOD)-Leucine-Rich Repeat (NLR) [8,9]. Caspases are known by alternative names, which have been in Table 1.
| S.No | Common name | Gene name/aliases |
|---|---|---|
| 1 | CASPASE 1 | Interleukin-1 [11] |
| 2 | CASPASE2 | ICH-1, NEDD-2 ( NEURAL Precursor Cell ExpressedDevelopmentally Downregulated 2 ) [12] |
| 3 | CASPASE3 | CPP32(cysteine protease protein)/Yama/apopain [13] |
| 4 | CASPASE4 | ICH-2 [14] |
| 5 | CASPASE5 | ICH-3 [15] |
| 6 | CASPASE6 | MCH-2(Melanin Concentrating receptor 2 hormone [16] |
| 7 | CASPASE7 | MCH3; CMH-1(CPP32 MCH2 homolog-1); LICE2(lymphocytic ICE- protease 3) [17] |
| 8 | CASPASE8 | CAP4(Catabolite Activator PROTEIN4)MACH; MCH5(Melanin- concentrating hormone, receptor5); FLICE(FADD-like IL-1beta converting enzyme) [18] |
| 9 | CASPASE9 | ICE-LAP6(ICE-like apoptotic protease6); MCH6(Melanin- Concentratinghormone 6); PPP1R569(Protein phosphatase 1, regulatory subunit 56.), APAF- 3(Apoptotic protease activatingfactor 1,) [19] |
| 10 | CASPASE10 | ALPS2(Autoimmune Lymphoproliferative syndromes2); FLICE2(Fas- associated death domain protein interleukin-1beta- converting enzyme 2); MCH4(mammalianCED3) homolog-4) [20] |
| 11 | CASPASE 11 | SIP1, CASP11; SFRS2IP; SRRP129; SRSF2IP [21] |
| 12 | CASPASE12 | CASP12 [22] |
| 13 | CASPASE14 | ARCI12 [23] |
Table 1: List of human caspases and their other alternate names.
Human Caspases: Caspases are classified on the bases of their role in apoptosis. (Casp 3, 6, 7, 8, and 9 in mammals), in inflammation (casp1, 4, 5, 12) in humans and casp 1, 11, and 12 in mice [10].
Model Organism
Model organism includes a set of species which are studied to understand human-related diseases (Welcome trust, human genome project) [24]. Model organisms are easy to maintain and breed in lab settings and have particular experimental advantages. Model organisms used for analysis of human genetic disorder. Yeast (Saccharomyces) and other model organism colonial slime mold (Dictyostellium discoideum) have been used to study the functions like metabolism, regulation of cell cycle, membrane targeting and even DNA repair. Simple invertebrate systems such as Drosophila, Caenorhabditis elegans are excellent models for examining the coordinated actions of genes that function as components of a common molecular machine such as a signal transduction pathway or a complex of physically interacting proteins. C. elegans has also been widely used for human-related studies as their genomes are 40% homologous.
Human disease gene homologs with Drosophila and show 75% of human disease genes are structurally related with the gene present in Drosophila and more than third of human gene are highly related to fruit fly [25].
Model organisms such as mouse, zebrafish, frog, and chicken seem to show some extent of relativity with the human system. Thus, they can be used to perform experimentation to understand the diseased state and thereby relevant treatment for the same.
Humans and chimpanzees share 98% of significant matches at the nucleotide level. It has been found that the protein-coding genes are showing high correlation. The genetic comparison is not simple due to the nature of gene repeats and mutations (Chimpanzee sequencing consortium) [26]. Chimpanzees have 48 chromosomes, two more than humans. It is thought that this is because, in a human ancestor, two pairs of chromosomes fused into a single pair.
| Scientific name of model organisms for caspase study | Common name | Reference |
|---|---|---|
| Danio rerio | Zebrafish casp3a,3b,6,7,2,8,9 | Zhu S, et al. Cancer Cell [27] |
| Xenopus laevis | African clawed toad casp 3,6,7,10,9,8 | Nakajima et al. [28], McCoy Francis et al. [29] |
| Mus musculus | Mouse casp 2,8,9,14,3,6,7 casp | Ashrafi S, et al. Neuron [30] |
| Pan troglodytes | Chimpanzee casp2,8,9,10,3,6,7 | Parsons MJ, et al. [31] |
| Bos taurus | Cattle casp3 ,6,7,2,8,9,10 | Hernandez LL, et al. J Endocrinol [32], Hoa Van Ba, Inho Hwang [33] |
| Lepus curpaeums | Rabbit casp3,6,7,2,8,10 | Lin HD, et al. RheumatolInt [34], Uniport [35] |
| Felis catus | Cat casp 3,6,7,2,8,9,10 | Taylor S, et al. DNA Seq [36], Ensembl [37] |
| Gallus gallus | Hen (chicken) casp2,3,6 | Kaiser CL, et al. Hear Res [38] |
Table 2:List of caspases present in other model organisms.
Methodology
Caspases of human and different model organisms were studied. In humans, a total of 14 caspases have been found. They are divided into three categories-
1. Initiator caspases
2. Executioner caspases
3. Inflammatory caspases
Caspase related specific amino acid and nucleotide sequences of each of these model organisms were retrieved from NCBI (National Center for Biotechnology Information).
We have performed sequence alignment to identify those model organisms which share a remarkable similarity with human caspases and human caspase receptors, in the process of evolution using the different software packages (Power Phylogenetic Web Repeater), T-coffee, phylogeny fr, and Muscle. The different model organisms (Gallus gallus, Feliscatus, Xenopuslaevis, Lepus curpaeums, Bos taurus, Daniorerio, Mus musculus, Pan troglodytes)
T-coffee is Tree-based Consistency Objective Function For alignment Evaluation. T-coffee is developed by Notrdame et.al. National Institute for Medical Research London. T-coffee is developed in ANSI C Programing language. This method is based on the popular progressive approach to multiple alignments [39].
Further to validate our result, we have used the software MUSCLE -Multiple Sequence Comparison by Log-Expectation. It was developed by Robert C Edgar et.al. This algorithm includes fast distance estimation using kmer counting (is n-gram of nucleic acid or amino acid sequences that can be used to identify certain regions within biomolecules like DNA (e.g. for gene prediction or proteins), progressive alignment using a new profile function, (Log expectation score- indicates how sensitively a likelihood function depends on its parameters), and refinement using tree dependent restricted partitioning [40].
In order to have a better understanding, we have performed phylogenetic analysis using PHYLOGENY.fr is developed by Dereeper et al. (CNRS France). The phylogeny.fr platform automatically performs identification of homologous sequences, their multiple alignments (muscle), the phylogenetic reconstruction (PhyML), and the graphical representation of the inferred tree (TreeDyn) [41]. Phylogenetic Web Repeater (POWER) has been used to perform phylogenetic analysis keeping in mind the concept of a molecular clock [42].
Results
The results generated from the bioinformatics-based software’s have been compiled in the form of different tables for human caspases 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 (Table 3).
| S.No | Human caspase name | Similarity matches using Tcoffee | Similarity matches using muscle |
|---|---|---|---|
| 1 | Casp 2 | Pan troglodytes | Pan troglodytes |
| Mus musculus | Lepuscurpaeums | ||
| Danio rerio | Mus musculus | ||
| 2 | Casp 8 | Pan troglodytes | Pan troglodytes |
| Feliscatus | Feliscatus | ||
| Bos taurus | Bos taurus | ||
| 3 | Casp 9 | Pan troglodytes | Pan troglodyte |
| Danio rerio | Feliscatus | ||
| Xenopus laevis | Mus musculus | ||
| 4 | Casp 10 | Pan troglodytes | Pan troglodytes |
| Feliscatus | Feliscatus | ||
| Lepus curpaeums | Lepus curpaeums | ||
| 5 | Casp 3 | Pan troglodytes | Pan troglodytes |
| Gallus gallus | Feliscatus | ||
| Feliscatus, Mus musculus | Lepus curpaeums | ||
| 6 | Casp 6 | Pan troglodytes | Pan troglodytes |
| Gallus gallus | Bos taurus | ||
| Danio rerio | Feliscatus | ||
| 7 | Casp 7 | Pan troglodytes | Pan troglodytes |
| Lepuscurpaeums | Bostaurus | ||
| Mus musculus | Feliscatus | ||
| 8 | Casp 1 | Bos taurus | Lepuscurpaeums |
| Drosophila melanogaster | Bos taurus | ||
| Gallus gallus | Mus musculus | ||
| 9 | Casp 4 | Pan troglodytes | Pan troglodytes |
| Mus musculus | Mus musculus | ||
| 10 | Casp 5 | Pan troglodytes | Pan troglodytes |
| Drosophila melanogaster |
Table 3: Human caspase matches with caspases of model organism shown by T-COFFEE and MUSCLE.
To understand the relationship of the model organism with human caspases, we have tried to study their ancestral relationship; these results of the software’s have been included in Table 4.
| S.no | Human caspase name | Close phylogenetic clade using phylogeny. fr | Close phylogenetic clade using power |
|---|---|---|---|
| 1 | Casp 2 | Pan troglodytes | Pan troglodytes |
| Mus musculus | Lepus curpaeums | ||
| Lepus curpaeums | - | ||
| 2 | Casp 8 | Pan troglodytes | No result |
| Feliscatus | |||
| Bos taurus | |||
| 3 | Casp 9 | Pan troglodytes | No result |
| Feliscatus | |||
| Mus musculus | |||
| 4 | Casp 10 | Pan troglodytes | No result |
| Lepus curpaeums | |||
| Bos taurus | |||
| 5 | Casp 3 | Pan troglodytes | No result |
| Feliscatus | |||
| Mus musculus | |||
| 6 | Casp 6 | Pan troglodytes | No result |
| Lepus curpaeums | |||
| Mus musculus | |||
| 7 | Casp 7 | Pan troglodytes | Pan troglodytes |
| Feliscatus | Feliscatus | ||
| Bos taurus | - | ||
| 8 | Casp 1 | Bos taurus | No result |
| Lepuscurpaeums | |||
| Mus musculus | |||
| 9 | Casp 4 | Pan troglodytes | Pan troglodytes |
| Mus musculus | Mus musculus | ||
| 10 | Casp 5 | Pan troglodytes | Pan troglodytes |
| Drosophila melanogaster | - |
Table 4: Phylogenetic analysis result has been summarized in tabular form using Phylogeny.Fr and POWER software respectively.
Discussion
The current study is related to similarities among the different caspases of model organism and human using bioinformatics approach. This has been achieved by identifying amino acid sequence level similarities and then performing phylogenetic association, which may direct specific model organism suitable for particular human caspases.
It has been revealed in our study that different executioner caspases, initiator caspases, and inflammatory caspases show a different level of association with the model organism under study Different animal models are studied in order to introduce a better drug in the market. Therefore, specific studies were conducted so that the level of drug efficacy could improve. In this study, an attempt has been made to determine that although there are different model organisms which are widely used these days, still not one model organism should be considered for studying all the caspases.
Our suggestion is for human caspases related organism, the perfect model organism is Pan troglodytes. The phylogenetic analysis tools indicate that the next level of similarity is seen in case of Feliscatus, in case of caspases 3, 7, 8, and caspase 9.
It has also been observed that inflammatory caspases may have adopted a different route of evolution. It has been seen that human caspase 1 shows a high level of similarity with Bos taurus and Lepuscurpaeums. This clearly indicates that we should rethink while performing experiments on Mus musculus, as it may not show the same level of association, thereby may lead to decrease in the efficacy of drug designed for humans and tested on the mouse.
Conclusion
The significance of apoptosis in neurodegenerative diseases, autoimmune diseases, cardiovascular diseases, and cancer has been widely acknowledged. Thus, identifying key molecules whose regulation can trigger or inhibit PCD is crucial. Investigating the molecular mechanisms of apoptosis regulation has revealed a number of promising therapeutic targets, which can be utilized in drug discovery and medical research. Any chemical/natural compound considered as a drug target, for delivery, will be tested on a model organism. The significance of this study increases as it indicates that different human caspases show variation in the relativity with other model organism caspases. This study is a stepping stone towards identifying the differences at the molecular level among the model organism considered for human caspases related studies. As it clearly indicates that human caspases show a different level of relativity with different model organisms, thereby Mus musculus may not be considered, rather Feliscatus, Bos taurus, and Lepus curpaeumsshows remarkable similarity after Pan troglodytes.
Our study supports the idea that caspase inhibitors/enhancers can be used as an effective means to control the treatment of certain diseases and similar types of experiments should be carried out in animal models where resemblance occur with the respective caspases and model organism.
References
- Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 2009; 284(33):21777-81.
- Li P, Nijhawan D, Budihardjo I, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91(4):479-89.
- Sakamaki K, Satou Y. Caspases: evolutionary aspects of their functions in vertebrates. J fish biol. 2009; 74(4):727-53.
- Chang DW, Xing Z, Pan Y, et al. c‐FLIPL is a dual function regulator for caspase‐8 activation and CD95‐mediated apoptosis. EMBO J. 2002;21(14):3704-14.
- Wu CC, Lee S, Malladi S, et al. The Apaf-1 apoptosome induces formation of caspase-9 homo-and heterodimers with distinct activities. Nat. Commun. 2016;7:13565.
- Wang J, Lenardo MJ. Roles of caspases in apoptosis, development, and cytokine maturation revealed by homozygous gene deficiencies. J Cell Sci. 2000;113(5):753-7.
- Alexei D, Michael B, Junying Y. A decade of caspases. Oncogene. 2003;22(53):8543-67.
- Nadiri A, Wolinski MK, Saleh M. The inflammatory caspases: key players in the host response to pathogenic invasion and sepsis. J Immunol. 2006;177(7):4239-45.
- Chai J, Shi Y. Apoptosome and inflammasome: conserved machineries for caspase activation. Natl Sci Rev. 2014; 1(1):101-18.
- Eckhart L, Ballaun C, Hermann M, et al. Identification of novel mammalian caspases reveals an important role of gene loss in shaping the human caspase repertoire. Mol Biol Evol. 2008;25(5):831-41.
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1499
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1503
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1504
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1505
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1506
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1507
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1508
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1509
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1511
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1500
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:10784
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:19004
- https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1502
- http://www.sanger.ac.uk/resources/downloads/human/
- Bier E, McGinnis W. Model organisms in the study of development and disease. Oxford Monographs On Medical Genetics. 2004;49:25-45.
- Sequencing TC, Waterson RH, Lander ES, et al. Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437(7055):69.
- Zhu S, Lee JS, Guo F, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer cell. 2012;21(3):362-73.
- Nakajima K, Takahashi A, Yaoita Y. Structure, expression, and function of the Xenopus laevis caspase family. J Biol Chem. 2000;275(14):10484-91.
- McCoy F, Darbandi R, Nutt LK. Methods for the study of caspase activation in the Xenopus laevis oocyte and egg extract. InCaspases, Paracaspases, and Metacaspases. Humana Press, New York, NY. 2014;119-40.
- Ashrafi S, Betley JN, Comer JD, et al. Neuronal Ig/Caspr recognition promotes the formation of axoaxonic synapses in mouse spinal cord. Neuron. 2014;81(1):120-9.
- Parsons MJ, Lester KJ, Barclay NL, et al. Replication of genome‐wide association studies (GWAS) loci for sleep in the British G1219 cohort. Am J Med Genet B Neuropsychiatr Genet. 2013;162(5):431-8.
- Hernandez LL, Limesand SW, Collier JL, et al. The bovine mammary gland expresses multiple functional isoforms of serotonin receptors. J Endocrinol. 2009;203(1):123-31.
- H. Van Ba, I. Hwang; Role of caspase‐9 in the effector caspases and genome expressions, and growth of bovine skeletal myoblasts . Dev Growth Differ. 2014;56(2):1440-690.
- http://www.uniprot.org/uniprot/?query=caspase+1%2C+rabbit+&sort=score
- Lin HD, He CQ, Luo QL, et al. The effect of low-level laser to apoptosis of chondrocyte and caspases expression, including caspase-8 and caspase-3 in rabbit surgery-induced model of knee osteoarthritis. Rheumatol Int. 2012;32(3):759-66.
- http://www.ensembl.org/Cat/Search/Results?q=caspase;site=ensembl;facet_feature_type=;fa cet_species=Cat
- Taylor S, Hanlon L, McGillivray C, et al. Cloning and sequencing of feline and canine ice-related cDNAs encoding hybrid caspase-1/caspase-13-like propeptides. DNA Seq. 2000;10(6):387-94.
- Kaiser CL, Chapman BJ, Guidi JL, et al. Comparison of activated caspase detection methods in the gentamicin-treated chick cochlea. Hear res. 2008;240(1-2):1-1.
- Notredame C, Higgins DG, Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment1. J of mol biol. 2000;302(1):205-17.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792-7.
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic acids res. 2008;36(2):465-9.
- Lin CY, Lin FK, Lin CH, et al. POWER: PhylOgenetic WEb Repeater-an integrated and user-optimized framework for biomolecular phylogenetic analysis. Nucleic acids res. 2005;33(2):553-6.