Original Article
, Volume: 10( 4)Effect of Age of Seeds on Rice Transformation with Drought Stress Inducible Transcription Factor Encoding Gene by Biolistic Method
Ibandalin M, Kishwar A and Aruna T*
Division of Biochemistry, Indian Agricultural Research Institute, India
- *Correspondence:
- Aruna T, Division of Biochemistry, Indian Agricultural Research Institute, New Delhi, India 110012 E-mail: bio@iari.res.in
Received: June 02, 2016; Accepted: June 20, 2016; Published: June 24, 2016
Citation: Ibandalin M, Kishwar A, Aruna T. Effect of age of seeds on rice transformation with drought stress inducible transcription factor encoding gene by biolistic method. Biochem Ind J. 2016; 10(4): 103.
Abstract
Embryogenic calli produced in vitro from dehusked mature seeds of rice were bombarded with a gene construct containing a drought inducible transcription factor encoding gene named as AP2/ERF-N22, driven under inducible promoter RD29A using Biolistic method. The bombarded calluses were selected on hygromycin containing selection medium. The transformation efficiency was found to be nil, 0.97% and 3.11% for overstored seeds ≥ a year, ≤ a year and fresh seeds respectively. The survival rate of plants was 87%. About 90% of the plants reached maturity and showed no negative phenotypic effects or aberrations as observed earlier under a constitutive promoter from that of the wild type.
Keywords
Calli; Rice; Mature seeds; Transformation; Bombardment; Transgenics; Regeneration; AP2/ERF
Introduction
Rice is one of the world’s most important crops and caters the food grain requirement of more than half of the world population. However, rice is relatively sensitive to salinity and water stress. In order to adapt and grow in different environments, plants have developed a complex signaling network. At the molecular level, various transcription factors (TFs) regulate expression of multiple stress response genes [1]. In the last few years various transcription factors have been characterized for their roles in plant stress responses. Among different TFs, AP2/ethylene-responsive element-binding protein (AP2/EREBP) family plays an important role in the regulation of diverse biological processes such as plant growth, development, and response to abiotic stresses [2,3].
To understand the regulatory networks and mechanisms of drought tolerance, researchers are manipulating activity of these genes in transgenic plants to ameliorate crop yield under stressful conditions. The literature studies have shown that overexpression under a constitutive promoter may have negative effect on the plants [4-6]. Earlier study of a differentially expressed AP2/ERF-N22 from drought tolerant Oryza sativa sp Indica cv N22 [7], when overexpressed in Arabidopsis under a constitutive promoter, caused phenotypic aberrations though it improved drought tolerance when compared to the WT [8].
Plant regeneration from embryo derived callus of rice, was first reported by Nishi et al. [9]. Since then, many methods using various explants such as protoplast, immature seeds and embryogenic tissues have been reported for efficient gene expression in rice [10]. Ge et al. [11] successfully standardized rice transformation by using another explants. Apart from the exploration of the use of various explants, researchers have studied various factors that affect and influence the transformation efficiency. Ramesh and Gupta [12] showed that time of incubation of callus prior to bombardment greatly affected the efficiency. Li et al. [13] reported a substantial increase in transformation efficiency after bombardment by excising the resistant calluses followed by continuous sub culturing during regeneration and plant growth. Mandal et al. [14] showed that the size and age of the callus before bombardment influences the rate of transformation in rice. Jadhav et al. [15] investigated the efficiency of transformation based upon the type of callus and induction medium. Partial desiccation of callus prior to and after bombardment influenced shoot and bud initiation and ultimately regeneration efficiency over the non-desiccated calluses irrespective of rice genotypes [16-18]. Age of the seeds has never been considered seriously as one of the factors that influences transformation. No matter how good the storage condition may be the viability of seeds decreases with time.
Taking this into account, we transformed rice using AP2/ERF family transcription factor by biolistic method as it is variety-independent, efficient and simple and also studied the effect of age of seeds on transformation efficiency. Further, overexpression of AP/ERF-N22 in rice was taken up under drought inducible promoter RD29A in this study to see whether negative phenotypic effects observed earlier in Arabidopsis under a constitutive promoter can be corrected [8].
Materials and methods
Designing of construct AP2/ERF-N22 (Accession no. EF362638) driven under CaMV35S promoter and Nos terminator was restricted out from the binary construct pBI121:CaMV35S-AP2-Nos (Figure 1a) and cloned in pCambia1200. The recombinant construct named as pC1200: CaMV35S-AP2-Nos (Figure 1c) was restricted to remove CaMV35S and replaced by inducible RD29A promoter to obtain recombinant binary construct, now named as pC1200: RD29A-AP2-Nos (Figure 1f).
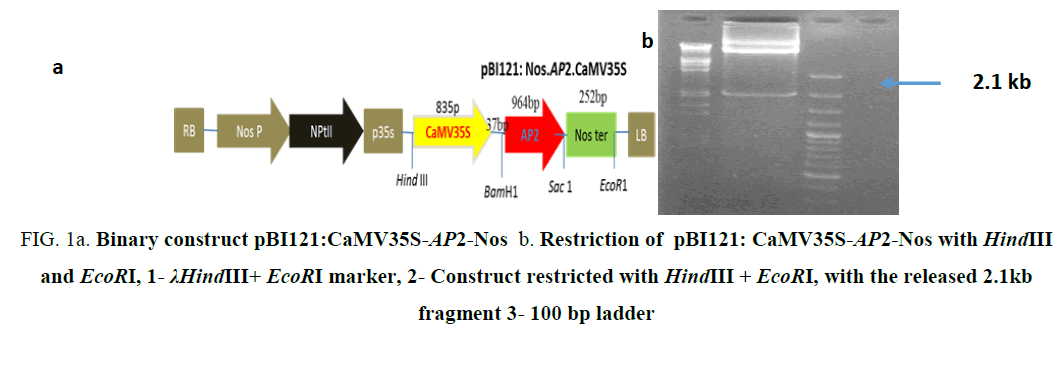
Figure 1a: Binary construct pBI121:CaMV35S-AP2-Nos b. Restriction of pBI121: CaMV35S-AP2-Nos with HindIII and EcoRI, 1- λHindIII+ EcoRI marker, 2- Construct restricted with HindIII + EcoRI, with the released 2.1kb fragment 3- 100 bp ladder
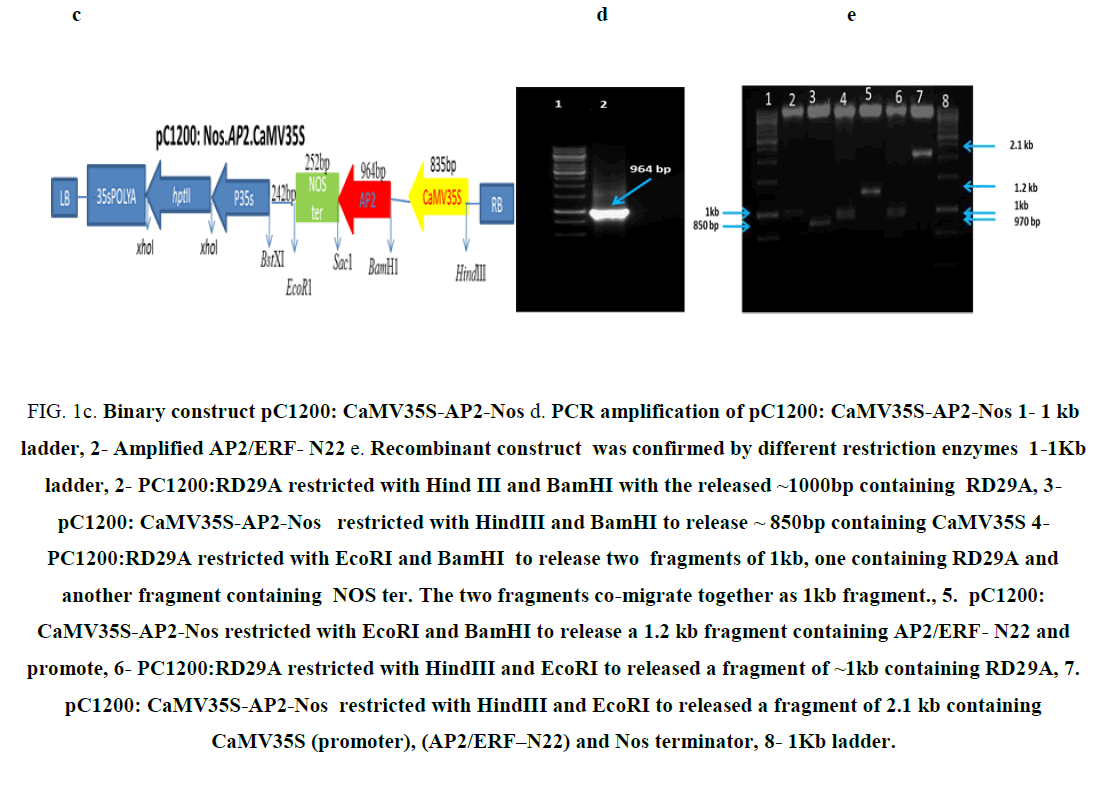
Figure 1c: Binary construct pC1200: CaMV35S-AP2-Nos d. PCR amplification of pC1200: CaMV35S-AP2-Nos 1- 1 kb ladder, 2- Amplified AP2/ERF- N22 e. Recombinant construct was confirmed by different restriction enzymes 1-1Kb ladder, 2- PC1200:RD29A restricted with Hind III and BamHI with the released ~1000bp containing RD29A, 3- pC1200: CaMV35S-AP2-Nos restricted with HindIII and BamHI to release ~ 850bp containing CaMV35S 4- PC1200:RD29A restricted with EcoRI and BamHI to release two fragments of 1kb, one containing RD29A and another fragment containing NOS ter. The two fragments co-migrate together as 1kb fragment., 5. pC1200: CaMV35S-AP2-Nos restricted with EcoRI and BamHI to release a 1.2 kb fragment containing AP2/ERF- N22 and promote, 6- PC1200:RD29A restricted with HindIII and EcoRI to released a fragment of ~1kb containing RD29A, 7. pC1200: CaMV35S-AP2-Nos restricted with HindIII and EcoRI to released a fragment of 2.1 kb containing CaMV35S (promoter), (AP2/ERF–N22) and Nos terminator, 8- 1Kb ladder.
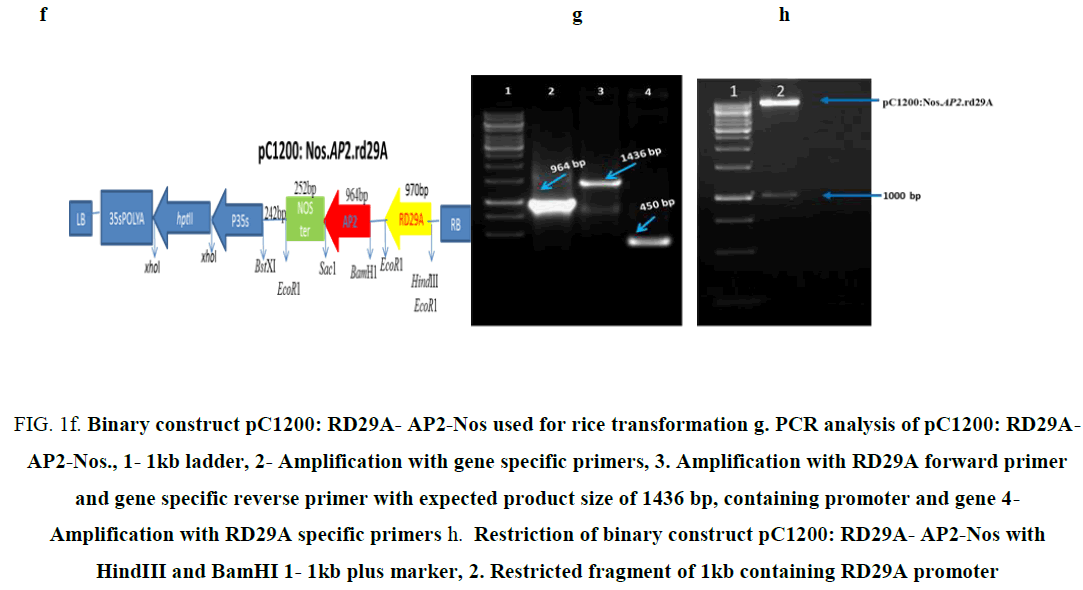
Figure 1f: Binary construct pC1200: RD29A- AP2-Nos used for rice transformation g. PCR analysis of pC1200: RD29A- AP2-Nos., 1- 1kb ladder, 2- Amplification with gene specific primers, 3. Amplification with RD29A forward primer and gene specific reverse primer with expected product size of 1436 bp, containing promoter and gene 4- Amplification with RD29A specific primers h. Restriction of binary construct pC1200: RD29A- AP2-Nos with HindIII and BamHI 1- 1kb plus marker, 2. Restricted fragment of 1kb containing RD29A promoter
Culture media and conditions
Culture media employed in these experiments were adjusted to pH 5.8 with NaOH. Callus induction medium (CIM) consisted of MS (Murashige and Skoog) powder with sucrose 30 g/l, Casein hydrolysate 200 mg/l, proline 500 mg/l, 2,4-D (1mg/ml) 2 ml/l and 2.5 g/l of phytagel. Osmotic media in addition to CIM consisted of sorbitol 0.2 M and manitol 0.2 M. Hygromycin selection media in addition to CIM consisted of 50mg/l hygromycin. Regeneration media in addition to CIM consisted of 300 mg/l glutamate, 6 benzylaminopurine (BAP) 2.5 ml/l, α napthalene acetic acid (NAA) (1 mg/ml) 0.75 ml/l, gelrite (in place of Phytagel) 3 g/l and hygromycin (50 mg/l). Solid rooting media composed of MS powder 2.2 g/l, sucrose 15 g/l, FeSO4 2.5 ml and gelrite 3 g/l. Liquid rooting media composition is same as solid rooting media with the exception of gelrite.
Experimental plant tissues
The dehusked mature seeds (200 to 500) of Pusa sugandh 2 were surface sterilized and inoculated into CIM (Figure 2a) for 1-2 weeks at 28°C under complete darkness. Nodular structures appeared from the general area of the scutellum. The callus was separated from the endosperm, cut into small pieces and arranged in clumps on osmotic medium plates as shown in Figure 2b. Prior to bombardment, these small calluses were incubated at 28°C under complete darkness for 4 h after which calluses were subjected to gold particle bombardment. After 48 h of incubation in dark at 28°C, each callus was transferred into selection media containing hygromycin (50 mg/l), (Figures 2c and 2d) and incubated at 28°C under complete darkness. After 15 days, the surviving calluses were transferred into fresh selection media. The rapidly proliferated calluses displayed somatic embryos in the form of whitish granular sectors. Some partially necrotic (brown sectors) and non-proliferating calluses were also observed, which were removed at each subculture. At the time of transfer, individual callusing embryos or callus pieces were broken using forceps into several small pieces which were maintained separately. During the subsequent two to three selection passages, callus pieces showing evidence of more vigorous growth were transferred to new selection plates. All callus tissue developed originally from each piece of embryo or callus was defined as single line. There were many plantlets obtained from each callus defined as a line.
Figure 2: a Explants (mature seeds of PusaSugandh 2) in CIM. Nodular structures appeared from the general area of the scutellum b Calluses on osmotic medium, ready for particle bombardment c&d Callus pieces on selection media e Callus on regeneration media f Regeneration of calluses into plantlets g Plantlets in rooting media with full development of primary and secondary roots h Plants in soilrite covered with white plastic bags for acclimatization. i Plants in green house
Microprojectile bombardment
All bombardments were conducted with biolistic Bio Rad PDS-1000/HeTM system. 30 mg of gold particles (0.6 μm) were washed with absolute alcohol thrice. 50 μl of 2.5 M CaCl2, 20 μl of 0.1 M spermidine and 10 μl of plasmid pC1200: RD29A-AP2-Nos (1 μg/ml) were added to the gold suspension of 10 μl, followed by 2 min incubation in ice, and short spin at 10000 rpm. The pellet was suspended in 90 μl ethanol, out of which 12 μl was evenly spread on macro carrier (cat#1652335, Bio Rad, India). Each experiment was bombarded at a pressure of 1100 psi. The distance from the stopping plate to the target was maintained 9 cm from the ruptured disc (cat#1652329, Bio Rad).
Molecular analysis
DNA was isolated using the CTAB method [19]. For analysis of transgenic plantlets, very small quantity of leaf (a part of leaf) sample from each line was taken with the help of sterile forceps. DNA was isolated by using micro pestle. However, for molecular analysis of fully grown mature plants, 500 mg of leaf tissue was taken. Total RNA was isolated from WT and transgenic plants under control and WDS. WDS was given by withholding the water from transgenics for three days. cDNA was synthesized. The integration of the gene was confirmed by PCR, Southern blot analysis and qRT -PCR. For PCR analysis specific primers for RD29A, hpt gene and AP2/ERF- N22 were used. PCR products of promoter and AP2/ERF-N22 were used as probes for hybridizing the blots. 5 μg DNA was digested with EcoRI. EcoRI sites are absent inside the gene and within the RD29A. Probe was labeled using the DecaLabel™ labeling kit (Fermentas, India) with 32P labeled ATP. Labeling of probe was done according to manufacturer’s protocol.
Result and Discussion
Designing of construct for transformation
The binary construct pBI121:CaMV35S-AP2-Nos (Figure 1a) carrying the gene AP2/ERF-N22 (Acccession no. EF362638) driven under CaMV35S promoter and containing kanamycin selection marker gene used earlier in our lab [8] was not used for rice transformation because of two reasons i.e. presence of constitutive CaMV35S (promoter) and kanamycin selection marker gene. The construct pBI121:Nos.AP2.CaMV35S (Figure 1a), carrying the gene AP2/ERF-N22 driven under CaMV35S promoter and kanamycin as selection marker was restricted with EcoRI and HindIII, to release the full casket of 2.1kb (Figure 1b) containing CaMV35S (promoter), (AP2/ERF-N22) and Nos terminator. Simultaneously, pCambia1200:RD29A was restricted with EcoRI and HindIII, to provide compatible sites for ligation. The released 2.1 kb fragment was ligated to the restricted pCambia1200:RD29A. The recombinant construct obtained was named as pC1200: Nos.AP2.CaMV35S (Figure 1c). Presence of gene was confirmed by PCR and restriction analysis. An amplicon size of 964 bp was obtained, corresponding to AP2/ERF–N22 (Figure 1d). Further confirmation was done by restricting the construct pC1200: Nos.AP2.CaMV35S with different restriction enzymes as shown in (Figure 1e). The recombinant pC1200: Nos.AP2.CaMV35S was restricted with EcoRI and BamHI to release a fragment of 1.2 kb containing AP2/ERF -N22gene + NOS ter (lane 5 in 1e ) and by restricting with HindIII and EcoRI to release a fragment of 2.1 kb containing promoter (CaMV35S) + gene (AP2/ERF- N22gene) + terminator (NOS ter) (lane 7 in 1e). The pC1200: Nos.AP2.CaMV35S was restricted with Hind III and BamHI to remove CaMV35S promoter (lane 3 Figure 1e). The RD29A was obtained from pCambia1200:RD29A by restricting with the same enzymes HindIII and BamHI (lane 2 Figure 1e) to generate compatible sites for ligation. The fragment RD29A was ligated to the restricted pC1200: Nos.AP2.CaMV35S, now lacking CaMV35S promoter. The recombinant binary construct DNA, now named as pC1200: Nos.AP2.rd29 (Figure 1f) was confirmed with PCR using gene specific primers as follows.
AP2/ERF-N22 Forward 5’ ATG GGA CAG TCG AAG AAG
AP2/ERF-N22 Reverse 5’ TCA GAT GAC GAG GCT AC
An expected amplicon size of 964 bp was obtained corresponding to AP2/ERF-N22 (lane 2 in Figure 1g)
Presence of inducble promoter RD29A was confirmed by using promoter specific primers
RD29A Forward 5’ ATT CGA ATG AGA AGG ATG TC
RD29A Reverse 5’ CCC TTT ATT CCT GAT CAT TG
As primers were designed from internal region an expected amplicon size of 450 bp was obtained (lane 4 in Figure 1g).
RD29A (forward) and AP2/ERF-N22 (reverse) primers were used to confirm the sense orientation of the gene with the promoter, using the following primers
RD29A Forward 5’ ATT CGA ATG AGA AGG ATG TC3’
AP2/ERF Reverse 5’ TCA CAC GCA GAA ATC CAC CTC CC 3’
An expected amplicon size of 1 kb was obtained (lane 3 in Figure 1g).Which compiles that the gene was cloned in the sense orientation and was driven under a constitutive promoter.
The construct was further confirmed by restriction digestion with HindIII and BamHI, which releases a desired fragment of ~1kb containing 970 bp (RD29A) (Figure 1h). The final construct under the name pC1200: Nos.AP2.rd29A was further confirmed by sequencing.
For rice transformation studies, hygromycin selection was preferred over kanamycin as it has been reported that protoplast-derived callus selected on kanamycin is very inefficient in terms of regeneration, and that a large number of albino plants arise from such experiments [20]. Hygromycin is more effective than kanamycin for transformation because rice shows natural resistance to kanamycin [21]. Also hygromycin neither inhibits regeneration, nor affects the fertility of transformant [22]. The hpt conferring resistance to hygromycin, has therefore been used as a marker gene in selection of transformants generated by both Agrobacterium-mediated and particle bombardment procedures [23]. Thus, binary vector pCambia1200 having hpt gene was taken for this study.
Effect of the age of seeds
Many studies have been done to identify suitable explants for in vitro callus induction. Various explants such as mature and immature embryos [24], leaf blade [25], mature seeds [26], microspore and anther [27,28], root [14,29] and coleoptile [17] have been used as good resources for in vitro production of embryogenic callus. Embryogenic calluses with high regeneration capacity are required for efficient rice transformation. Though immature tissues, which contain a large number of meristematic cells are more suitable for production of embryogenic callus as compared with mature seeds, especially for recalcitrant genotypes, but preparation of immature embryos on a large scale is tedious and has seasonal limitations [30]. Whereas, mature seeds have the advantage of convenient storability and ready availability throughout the year. Further using mature seeds makes the protocol for rice transformation efficient and simple for easier production of improved genotypes by genetic engineering with the potential for gene function studies.
It was observed that seeds (explants) which were stored for a long time showed delay in callus formation, or no callus formation at all, while some showed delayed regeneration or no regeneration at all (Table 1). However, when fresh seeds (1-2 weeks after harvesting) were used there was neither a delay in callus formation, nor in regeneration (Table 1). Further, it was observed that fresh seeds gave rise to healthy callus, which started to show initial greening, with regeneration within a period of one month (Figure 2e). These were transferred into bottles, to allow them to regenerate further. Four weeks after transferring into the regeneration medium and incubation under photoperiod (16h light/8 h dark) in growth room at 25°C, somatic embryos rapidly regenerated into shootlets with small roots (Figure 2f). Each plantlet was separated and transferred into solid rooting media for enhancing full root growth. After a week or so, with full development of primary and secondary roots, each plantlet was then transferred into a liquid rooting media which lead to development of normal roots (Figure 2g). Each plantlet confirmed to be transgenic was transferred into small pots containing soilrite and covered using transparent plastic bags for acclimatizing the plants. To allow air exchange, small holes were made in plastic bags (Figure 2h). After 7-12 days, the plastic bags were removed and plantlets were transferred into pots and allowed to grow in greenhouse till maturity (Figure 2i). This study indicates that the ability of callus formation is largely dependent on the age of the seeds which in turn is a prerequisite for highly efficient rice transformation.
| A. Transformation efficiency of this experiment when old stored seeds were used | |||||||||||
| Expt | %of CIM | % calli bombarded | % calli selected | Time of regeneration | % of regeneration | Total no. of plantlets | Total positive plants | Plants survived after transplant | TE* (%) | TE* (%) mean | |
| 1. | 9.78 | 100 | 83.33 | 5 months | NT | NT | NT | NT | NT | ||
| 25.7 | 100 | 86.67 | 4-5months | NT | NT | NT | NT | NT | 0.95 | ||
| 56.8 | 64.44 | 45.52 | 4months | 100 | 284 | 8 | 4 | 2.8 | |||
| B. Transformation efficiency of this experiment when freshly harvested seeds were used. | |||||||||||
| Expt. | %of CIM | % calli bombarded | %calli selected | Time of regeneration | % of regeneration | Total no. of plantlets | Total positive plants | Plants survived after transplant | TE* (%) | TE* (%) mean | |
| 2. | 39.25 | 96 | 45.83 | 1month | 90 | 187 | 6 | 6 | 2.5 | ||
| 3. | 52.96 | 99.65 | 26.32 | 1month | 100 | 359 | 7 | 7 | 1.95 | 2.93 | |
| 4. | 77.72 | 100 | 33.33 | 1month | 100 | 465 | 13 | 13 | 4.33 | ||
| C. Total transformation efficiency. | |||||||||||
| Expt. | %of CIM | % calli bombarded | %calli selected | Time of regeneration | % of regeneration | Total no. of plantlets | Total positive plants | Plants survived after transplant | Survival rate | TE* (%) | TE* (%) mean |
| 1. | 9.78 | 100 | 83.33 | 5 months | - | - | - | - | - | ||
| 25.7 | 100 | 86.67 | 4-5months | - | - | - | - | - | |||
| 56.8 | 64.44 | 45.52 | 4 months | 100 | 284 | 8 | 4 | 87 % | 2.8 | 1.93 | |
| 2. | 39.25 | 96 | 45.83 | 1 month | 90 | 187 | 7 | 7 | 2.5 | ||
| 3. | 52.96 | 99.65 | 26.32 | 1 month | 100 | 359 | 6 | 6 | 1.95 | ||
| 4. | 77.72 | 100 | 33.33 | 1 month | 100 | 465 | 13 | 13 | 4.33 | ||
The transformation efficiency (% TE) which is obtained by calculating the ratio of the (No. of positive plantlets/ No. of calli bombarded)*100.
The percentage of calli (% of CIM) obtained which is calculated as total (No. of calli obtained to the total no. of seeds inoculated)*100, calculated values are listed in the table.
The percentage of regeneration (% regeneration) is calculated as the ratio of the (No. of regenerated calli to the No. of calli obtained after selection)*100.
The survival rate (%) is calculated as the ratio (total no. of positive plants survived/ total no. of positive plants obtained)* 100.
The calculated values for each experiment are shown in the table.
Table 1: Survival rate and transformation efficiency of plants.
Transformation efficiency and survival rate of plants
The protocol which we used for biolistic transformation [31] has already reported transformation efficiency of 87% with the same genotype. Our objective was not to increase or analyze transformation efficiency. Since age of the seeds has never been considered as one of the factors that influences transformation, we correlated age of seeds with regeneration time and transformation efficiency. Transformation efficiency and regeneration time are greatly influenced by the age of seeds in this protocol. In this experiment the standardization of the protocol was based on the use of stored seeds and freshly harvested seeds. The transformation efficiency with stored seeds was 0.95%, with delayed regeneration time (about 4 to 5 months) and some callus did not regenerate into plantlets. However freshly harvested seeds (1 week after harvesting) showed a marked increase in the transformation efficiency (2.93%) and callus regenerated into number of plantlets over a period of 1month (Table 1).
From this experiment we can observe that selection of explants greatly influence the transformation efficiency. The overall transformation efficiency from the present experiment was found to be 1.93%. Apart from considering the age of seeds as an influential factor for enhancing the transformation efficiency there are other factors too. As seen in previous studies, the procedure used for precipitation of DNA onto gold micro-particles prior to biolistic transformation may impact transformation success and efficiency [32,33]. Typically, DNA is usually precipitated onto micro-particles in the presence of spermidine and CaCl2 for biolistic transformation of plants [33], which was used in the present experiment. Sivamani et al [33] reported that using a cationic polymer protamine instead of spermidine for biolistic transfer of full plasmids to rice and maize resulted in over a five-fold increase in transient transformation efficiency and a 3.3-fold increase in stable transformation efficiency. Stable transformation of sugarcane, by biolistic transfer using protamine, spermidine and Seashell protocols were equally effective [34]. Interestingly Sivamani and group in their observations, showed spermidine resulted in significantly higher transient expression than the other two precipitation protocols. Xiong et al. [35] however, found that protamine protects plasmid DNA for a longer period from DNAase degradation than spermidine. Various studies have shown different ways to increase the transformation efficiency. Thus no single protocol is said to be the best, as it depends on many factors which need to be studied.
Rate of regeneration is greatly influenced, by the quality of seeds and is very important for successful tissue culture. Survival rate of plants was 87%, after their transfer to soil in the greenhouse and more than 90% of plants reached maturity. Most T0 plants had normal morphology and were self-fertile.
Conclusion
Transgenic lines of different crop plants have been successfully obtained by particle bombardment method. Agrobacterium-mediated transformation systems have also been used successfully in recent years for developing transgenic rice [36] maize [37] barley [38] and wheat [39]. However, particle bombardment remains preferred method for developing transgenics of monocot species, because at least in rice it is variety-independent, efficient and simple. In our experiments we have found age of seed to be an important factor that influenced the rate of regeneration and transformation efficiency. The transformation efficiency was found to be nil, 0.97% and 3.11% for overstored seeds ≥ a year, ≤ a year and fresh seeds respectively. The survival rate of plants was 87%. About 90% of the plants reached maturity and showed no negative phenotypic effects or aberrations as observed earlier in Arabiopsis under a constitutive promoter. Several features of the transformation protocol reported in this study will facilitate its adoption by other laboratories.
Acknowledgements
The authors acknowledge Dr. T Mohapatra, Director IARI, New Delhi for binary vector pCambia1200: RD29A and allowing use of particle gene gun in NRCPB. The authors thankfully acknowledge National Phytotron Facility to carry out the various experiments and the financial support from SERB, DST.
References
- Ali K, Rai RD, Tyagi A. Expression analysis of bZIP transcription factor encoding genes in response to water deficit stress in rice.Ind J Exp Biol. 2016;54:332-7.
- Mawlong I, Ali K, Tyagi A. Cloning and characterization of a water deficit stress responsive transcription factor gene from Oryza sativa L. Ind J Exp Biol. 2016;54(1):26-36.
- Abogadallah GM, Nada RM, Malinowski R, et al. Overexpression of HARDY, an AP2/ERF gene from Arabidopsis, improves drought and salt tolerance by reducing transpiration and sodium uptake in transgenic Trifoliumalexandrinum L. Planta.2011;233(6):1265-76.
- Morran S, Omid E, Tatiana P, et al. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB⁄CBF factors. Plant Biotech J. 2011;9(2):230-49.
- Gargul JM, Mibus H, Serek M. Constitutive overexpression of Nicotiana GA2ox leads to compact phenotypes and delayed flowering in Kalanchoe¨blossfeldiana and Petunia hybrid. Plant Cell Tiss Org. 2013;115(3):407-18.
- Atkinson NJ, Lilley CJ, Urwin PE. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013;162(4):2028-41.
- Mawlong I, Ali K, Kurup D, et al. Isolation and characterization of an AP2/ERF-type drought stress inducible transcription factor encoding gene from rice. J Plant Biochem Biotech. 2014;23(1):42-51.
- Mawlong I, Ali K, Srinivasan R, et al. Functional validation of a drought-responsive AP2/ERF family transcription factor-encoding gene in Arabidopsis. Mol Breed. 2015;35:1-14.
- Nishi T, Yamada Y, Takahashi E. Organ re-differentiation and plant restoration in rice callus. Nature. 1968;219(5153):508-9.
- Cantrell RP, Reeves TG. The cereal of the world’s poor takes center stage. Science.2002;296:53.
- Ge X, Chu Z, Lin Y, et al. Tissue culture system for different germplasms of indica rice. Plant Cell Rep. 2006;25(5):392-402.
- Ramesh M, Gupta AK. Transient expression of glucuronidase gene in indica and japonica rice (Oryza sativa L.) callus cultures after different stages of co-bombardment.Afr J Biotechnol. 2005;4(7):596-600.
- Li L, Qu R, Kochko A, et al. An improved rice transformation method using the biolistic method. Plant Cell Rep. 1993;12(5):250-55.
- Mandal AB, Maiti A, Biswas A. Somatic embryogenesis in root derived callus ofindica rice. Plant Tiss Cult.2003;13(2):125-33.
- Jadhav PV, Dudhare M, Taranjeet S, et al. Assessment of critical factors influencing callus induction, in vitro regeneration and selection of bombarded indica rice genotypes. J Cell Plant Sci. 2011;2(2):24-42.
- Diah A, Bhalla PL. Plant regeneration from mature embryo-derived callus of Australian rice (Oryza sativa L.) varieties. Aust J Agric Res.2000;51:305-12.
- Chand S, Sahrawat AK. Stimulatory effect of partial desiccation on plant regeneration in indica rice (Oryza sativa L.). J Plant BiochemBiotechnol. 2001;10(1):43-47.
- Saharan V, Yadav RC,Yadav NR, et al. High frequency plant regeneration from desiccated calli of indica rice (Oryza Sativa l.).Afr J Biotechnol. 2004;3(5):256-9.
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed.New York: Cold Spring Harbor Laboratory Press;1989.
- Toriyama K, Arimoto Y, Uchimiya H, et al. Transgenic rice plants after direct gene transfer into protoplasts.Biotechnol. 1988;6:1072-74.
- Christou P, Ford TL. Parameters influencing stable transformation in rice embyrogenic tissue culture and recovery of transgenic plants using electric discharge particle acceleration. Ann. Bot. 1995;75:407-13.
- Aldemita RR, Hodges TK. Agrobacterium tumefaciens-mediated transformation of japonica and indica rice varieties. Planta.1996;199:612-17.
- van den Elzen PJ, Townsend J, Lee KY, et al. Chimeric hygromycin resistance gene as a selectable marker in plant cells, Plant Mol. Biol. 1985;5(5):299-302.
- Lee SY, Kin HS, Kwon TO. Variation in anther culture response and fertility of backcrossed hybrids between indica and japonica rice (Oryza sativa). Plant Cell Tiss Org.2004;79:25-30.
- Yan CJ, Zhao QH. Callus induction and plantlet regeneration from leaf blade of Oryza sativa L. subsp. Indica. Plant SciLett. 1982;29:175-82.
- Sivamani E, Shen P, Opalka N, et al. Selection of large quantities of embryogeniccalli from indica rice seeds for production of fertile transgenic plants using the biolistic method. Plant Cell Rep.1996;15(5):322-27.
- Lentini Z, Reyes P, Martinez CP,et al. Androgenesis of highly recalcitrant rice genotypes with maltose and silver nitrate. Plant Sci. 1995;110,127-38.
- Lee KS, Jeon HS, Kim MY. Optimization of a mature embryo based in vitro culture system for high-frequency somatic embryogenic callus induction and plant regeneration from japonica rice cultivars. Plant Cell Tiss Org, 2002;71:9-13.
- Sticklen MB. Direct embryogenesis and fertile plants from rice root culture. J Plant Physiol. 1991:138:577-80.
- Morrish F, Vasil V, Vasil IK. Developmental morphogenesis and genetic manipulation in tissue and cell cultures of the gramineae. Adv Genet.1987;24, 432-99.
- Latha GM, Raman KV, Kumar S, et al. High frequency regeneration and transformation of indica rice variety Pusa Sugandh2. ORYZA.2014;51:24-33.
- Rasco GS, Riley A, Barcelo P, et al. Analysis of particle bombardment parameters to optimiseDNA delivery into wheat tissues. Plant Cell Rep. 1999;19:118-27.
- Sivamani E, DeLong RK, Qu R. Protamine-mediated DNA coating remarkably improves bombardment transformation efficiency in plant cells. Plant Cell Rep. 2009;28(2):213-21.
- Allahi S, Khodaparast SA. Mohammad MS. Agrobacterium-Mediated Transformation of Indica Rice: A Non-Tissue Culture Approach. Int. J AgrInnov Res.2014;3(2):467-72.
- Xiong AS, Jiang HH, Zhuang J, et al. Expression and function of a modified AP2/ERF transcription factor from Brassica napus enhances cold tolerance in transgenic Arabidopsis. MolBiotechnol.2013;53(2):198-206.
- Wang Y, Zeng H, Zhou X, et al. Transformation of rice with large maize genomic DNA fragments containing high content repetitive sequences. Plant Cell Rep.2015;34(6):1049-61.
- Cao SL, Masilamany P, Li WB, et al. Agrobacterium tumefaciens mediated transformation of corn (Zea mays L.) multiple shoots. Biotech Biotechnol Eq.2014;28(2):208-16.
- Harwood W A, Bartlett J, Alves S, et al. Barley transformation using Agrobacterium-mediated techniques. In: HuwD Jones and Peter R Shewry (eds). Methods in Molecular Biology, Transgenic Wheat, Barley and Oats. 2009; Vol 478, Chapter 9, pp137-147.
- Sparks CA, Doherty A, Jones HD. Genetic transformation of wheat via Agrobacterium-mediated DNA delivery, Methods.Mol Biol.2015;1099:235-250.