Original Article
, Volume: 2( 1)Advances in the Treatment of Primary Liver Cancer
- *Correspondence:
- Ashraf G, Department of Biology, Research Institute for Fundamental Sciences, University of Tabriz, Tabriz, Iran, Tel: 98-411-33393924, E-mail: aghz_bioch@yahoo.co.in
Received: June 24, 2016; Accepted: June 29, 2016; Published: July 07, 2016
Citation: Ashraf G. Real Time Based RT-PCR Detection of DUF538 Gene Expression in Drought-Challenged Celosia. Biochem Mol Biol 2016; 2(1): 101
Abstract
Primary liver cancer (PLC) is one of the common malignant tumors. With high incidence and poor prognosis, PLC has become one of the major diseases which is seriously harmful to human life and health. In recent years, the research work have been summarized on the status of the treatment of primary liver cancer, and great progress has been made, especially the significant progress in the early diagnosis, surgical treatment, and comprehensive treatment of liver cancer made to improve the quality of life of patients. In spite of many methods can be used to improve the quality of life, there are still issues need to be studied deeply. The overview of the progress in the therapy research of liver cancer had been done in this paper that may provide assistance for the further treatment of such disease.
Keywords
Celosia; Drought stress; DUF538; Gene expression; Real-Time PCR
Introduction
DUF538 protein superfamily consists of several plant proteins of unknown functions. DUF538 is the only significant and recognizable conserved domain that has been reported on this protein superfamily, thus far. The protein members of this superfamily have been distributed in wide ranges of monocotyledonous and dicotyledonous plant species [1,2]. They have been estimated to have molecular weights of about 19 kDa to 21 kDa encoding around 170 amino acids. Recently, the three dimensional petal-like structure of Arabidopsis thaliana DUF538 protein has been determined by NMR and released to the universal protein databases (PDB ID: D1ydua1). It has been shown to possess a protein structure dominated by β-strands.
Several hundreds of DUF538 deduced amino acid sequences have been deposited as hypothetical proteins in universal databases. But, none of the members of this protein superfamily has been found in algae and cyanobacteria so far [2]. DUF538 domain containing proteins have been mostly identified using genome annotation tools and also been cloned as inducible genes from plants challenged with various environmental stresses such as nutrient deficiency (GenBank, BFT415717), crown gall (GenBank, BG131101), and mixed elicitors (GenBank, AW040635) from tomato, and mild drought from Populus (GenBank, CU232042) and Celosia plants (GenBank, AJ535713) without real time analysis and comparison of its expression level [3]. On the basis of the high phosphorylation potential, DUF538 proteins have been predicted to play important regulatory roles in different stress-challenged plants [4]. Our report has revealed that the exogenously applied fusion form of a DUF538 protein by using a plant tissue abrading material activates the redox system of the plant cells [1].
In a recent study, DUF538 proteins have been suggested to affect the bacterial growth rates through the binding to the lipopolysaccharide molecules on the outer leaflet of the bacterial membranes as like as bactericidal permeability increasing proteins in mammalians innate immune system [5]. Then after, by using the bioinformatics tools, DUF538 tertiary structures was predicted to be similar to the esterase-type hydrolases or lipolytic enzymes of which carboxy esterease type B, acyl-peptide hydrolase, entrochelin esterease, and peroxisomal long chain acyl-coA hydrolase were the best matches [6].
Keeping all this information in view, DUF538 members may be suggested as a common stress-related strategy in plant system. In the present research work, as a part of our studies to increase our knowledge regarding the possible roles of DUF538 protein superfamily in plant system, our attempt was made to investigate the expression profile of DUF538 gene expression in the leaf tissues of Celosia cristata plants challenged with drought stress.
Materials and Methods
Chemicals
Trizol reagent (Cat. no. RN7713C; RNX™; CinnaGen) was used for total RNA isolation, mRNA purification kit was provided by QIAGEN, USA (Cat. No.70022). AcessQuickTM RT-PCR System for cDNA synthesis was purchased from Promega (Cat. no. A1701). SYBR Green dye used for real-time detection of gene expression was from Bio Rad Company. Fermentas DNA extraction kit (Cat. no. K0513) was used for the purification of the PCR product from the agarose gel. Plasmid vector pGEM-T easy (Cat. no. A1360; Promega) was used for PCR product cloning. All of the other chemicals used in this research work were of molecular biology grades.
Plant materials
The seeds of Celosia cristata were taken from our Laboratory stock. Test plants were allowed to grow under normal laboratory conditions of day to night period and humidity. In order to collect the leaf materials, test plant was well watered until sixth leaf stage and then after the water was withheld till the plant was visibly welted. Drought stress conditions were continued for 4 weeks. Experimental materials were collected from leaf tissues before and after stress treatment.
Total RNA isolation and mRNA purification
Total cellular RNA was separately isolated from test materials using Trizol reagent, separately. About 0.2 g of each material was fine powdered using liquid N2 and 2 ml of Trizol reagent was added to homogenize it at room temperature (RT). 200 μl of chloroform was added to the mixture, mixed for 15 seconds, incubated on ice for 5 min and centrifuged at 13000 rpm for 15 min. The upper phase was transferred to another tube and RNA was precipitated with an equal volume of isopropanol. The pellet was washed in 1 ml of 75% ethanol, dried at RT and dissolved in 30 μl RNase-free water. The integrity of the RNA was tested on 1% non-denaturing agarose gel using TBE running buffer. Poly (A+) RNA was purified from total RNA using oligo dT-columns according to the provided kit protocol. The integrity of the purified mRNA was also analyzed by electrophoresis using 1% non-denaturing agarose gel. The quantity of the RNA in the starting materials for the next experiments was measured spectrophotometrically [7].
Primer designing and cDNA synthesis
Specific primers used for the amplification were designed based on already reported DUF538 transcript sequence [3]. The primer sequences were designed by online internet-based Primer3 software.
In order to analyze the expression pattern of DUF538 gene, the RT-PCR reactions were separately performed using one-step AcessQuick™ RT-PCR System. About 0.5 μg of each mRNA sample was mixed with 25 μl Master Mix (2X) and 1 μl of correspondent primer set. The mixtures were adjusted to a final volume of 50 μl using nuclease-free water. The reaction mixtures were incubated at 45°C for 45 min and preceded with PCR cycling. PCR was carried out after a pre-denaturation stage at 95°C for 3 min in 25 cycles. The nucleotide sequence of the used primer set and the details of PCR steps were performed as our previous work [1] and presented in Table 1.
| PremierSequences | |
| Left Premier:5 ‘ C G A C C T T C A T T A A T G G A C C 3’ | |
| Right Premier: 5 ‘ A C T C A T C C A A G C T C G C A A A 3’ | |
| PCR amplification Steps | |
| Denaturation | 95\C / 1 min |
| Annealing | 55\C /1 min |
| Extension | 72\C /1.5 min |
| Final Extension | 72\C /10 min |
Table 1: Premier set & PCR amplification Steps.
In the next step, the amplified products were extracted from the agarose gel, cloned in pGEM-T easy cloning vector [7]. The cloned fragments proceeded for the sequencing in Microsynth DNA sequencing center at Switzerland.
Expression analysis by real-time reverse transcription PCR
The expression level of the DUF538 transcripts in test samples were analyzed by real-time RT-PCR using iQ SYBR Green supermix (Bio Rad) on Miniopticon Real Time PCR Detection System (Bio Rad). Experiments were performed in three replicates and the mean values of the data were presented. All the expression data were normalized by adjusting the expression level of actin gene in test samples and the relative fold expression was calculated using the 2-ΔΔCt value [8].
Sequence analysis of the isolated fragments
The nucleotide sequences of the amplified cDNAs were analyzed by computing at BLAST (Basic Local Alignment Search Tool) at http://www.ncbi.nlm.blast.com/, and CLASTALW sequence alignment software at http://www.genome.jp/ and Expasy proteomic tools at http://www.expasy.org/tools/.
Results and Discussion
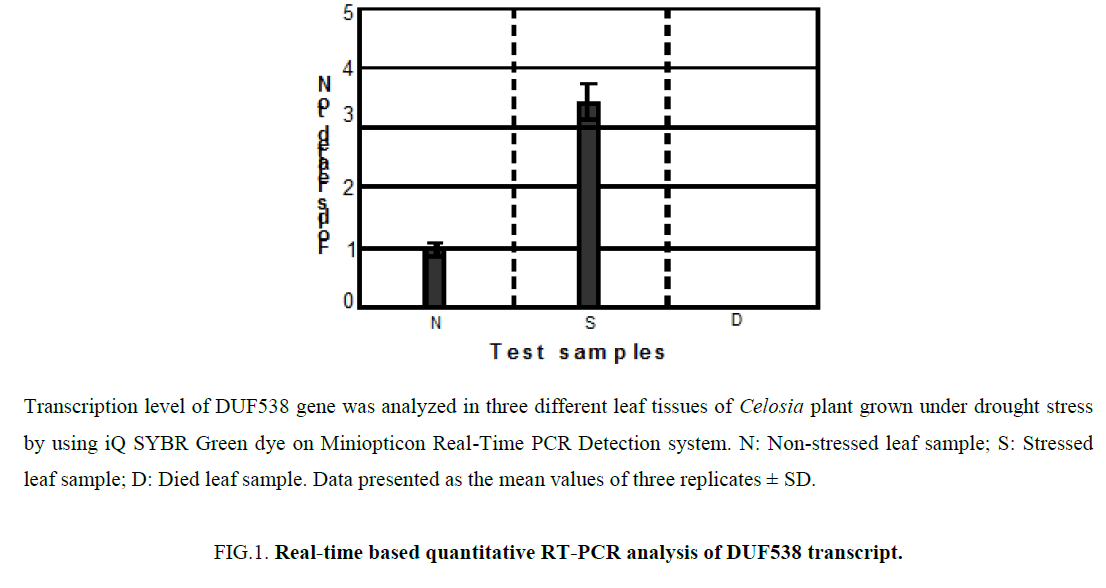
In the previous work, we identified a drought stress inducible DUF538 gene from the leaves of Celosia cristata plants [3]. Analysis of RT-PCR end products on 1% agarose gel revealed that the expression of DUF538 transcript is consistently detectable in the leaf material of non-stressed and stressed samples. Only the sample collected from died leaf tissues failed to show evidence of DUF538 gene expression on agarose gel. Herein, we extended our expression studies by real time analysis on drought challenged leaves of Celosia plant. To prime the expressed DUF538 gene from leaf tissues, previously reported primer pair for the Celosia DUF538 was used [3]. The presence and the level of DUF538 transcript expression were detected by real time RT-PCR method using the test samples collected from leaf tissues at three time points including non-stressed, stressed and died. Figure 1 shows the expression and quantification of the relative fold change in the expression level of DUF538 gene in three different leaf samples. All data were normalized and related to L0 (non-stressed leaf sample considered as control) using 2-ΔΔCt value [8]. Like to the previous results obtained by agarose gel, analysis of the 2-ΔΔCt values reliably revealed that Celosia DUF538 gene is expressed in both stressed and non-stressed leaf test samples except for dried leaf material. Results showed that the expression level of DUF538 varies in stressed and non-stressed samples. It was shown that the expression level of DUF538 gene is considerably higher in stress challenged tissues (Figure 1). In comparison with non-stressed tissues, there is about 3.2-3.6 folds’ increase in stressed leaf samples. The expression level of DUF538 was found to be no detectable by real time expression analysis as by agarose gel in dried leaf samples. The results indicated that the expression level of DUF538 gene is increased in stressed leaf of Celosia plant.
Figure 1: Real-time based quantitative RT-PCR analysis of DUF538 transcript.
Transcription level of DUF538 gene was analyzed in three different leaf tissues of Celosia plant grown under drought stress by using iQ SYBR Green dye on Miniopticon Real-Time PCR Detection system. N: Non-stressed leaf sample; S: Stressed leaf sample; D: Died leaf sample. Data presented as the mean values of three replicates ± SD.
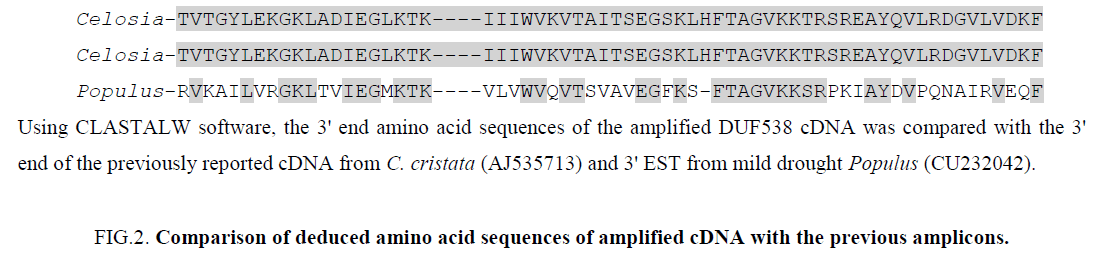
In order to analyze the sequences of the stressed and non-stressed amplification products, they were cloned and sequenced. The nucleotide sequences comparison analysis results revealed that the amplified cDNA from stressed and non-stressed tissues were 100% identical (nucleotide sequence data not presented). Comparison of their deduced amino acid sequences by CLASTALW software indicated that the amino acid sequences of amplified cDNAs are 100% and 50% identical to the previously reported DUF538 cDNA from Celosia plant and EST from Populus [3] (Figure 2).
The expression of DUF538 gene by basic RT-PCR method had been previously reported from different plants grown under stressful environmental conditions such as nutrient deficiency (GenBank, BFT415717), crown gall (GenBank, BG131101), and mixed elicitors (GenBank, AW040635, and mild drought from Populus (GenBank, CU232042) and Celosia plants (GenBank, AJ535713) [3]. All these expression data were to our knowledge without real time analysis and comparison of gene expression level. The present real-time data related to DUF538 gene expression suggest that DUF538 protein members are highly induced under drought condition and might have potential role in drought-stressed plants such as Celosia. It has been understood that the products of drought stress inducible genes are classified into functional and regulatory proteins. The regulatory proteins regulate the various environmental stress signal transduction pathways. They have been usually known to control the expression of other stress-inducible genes in plant system [9-11]. These include various transcription factors, protein kinases, protein phosphatases, enzymes involved in phospholipids metabolism and other signaling molecules such as calmodulin – binding protein [12,13]. According to the constitutive as well as stress-induced gene expression of DUF538 protein member in Celosia plants and considering its expression under other stress-challenged plants, DUF538 protein superfamily seem to be putative candidate for common stress-related proteins possibly common regulatory factor which may be involved in various plant stress responses.
The gene expression profiles of plants exposed to drought stress have been previously analyzed by the microarray technology and other molecular and genomics methods particularly in Arabidopsis and rice plants. Chaperone molecules, late embryogenesis abundant proteins, osmotics, osmolyte biosynthesizing enzymes, antifreeze proteins, mRNA-binding proteins, water channel proteins, sugar and amino acid transporters, detoxifying enzymes, anti-oxidative proteins and various hydrolytic enzymes such as proteases are the well-known drought-stress related elements [14,15]. The high similarity score between DUF538 family members and hydrolase enzymes [5] may be a potent clue for the drought-stress related biological activity of DUF538 domain containing proteins in plants. This proposal needs for further clarification in the future.
The present real-time data may suggest that DUF538 gene activity may be as one of the drought stress defense responses in plant system. Our survey on the distribution of DUF538 members by using internet-based ortholog cluster analysis showed that DUF538 superfamily is distributed widely in dicot and monocot plants (Figure 3). Basal magnoliophyta, ferns and mosses were also found to have got this protein. But, animals, green algae, red algae, fungi, protests and prokaryotes were not found to contain DUF538 proteins. The distribution pattern of DUF538 in eukaryotic photosynthesizing organisms implies that the functional properties of this protein superfamily might be conserved and it might play an important role in photosynthetic organisms. On the other hand, the recent nomination of DUF538 protein family as the potential structural and functional homologue of mammalians bactericidal protein in plants provide a basis to study the potent anti-stress functions of this protein family or their homologues in mammals, too [5].
Figure 3: Analysis of DUF538 domain distribution in nature. Sequence organisms include Eudicots, Monocots, Basal Magnoliophyta, Ferns and Mosses.
As an initial report, we hope our results will provide a new research gate to study the detailed bio-physiological roles of DUF538 gene product in plant or animal system with regard to environmental stress stimuli.
Acknowledgements
The author of this study is thankful to Research Institute for Fundamental Sciences, University of Tabriz, Iran for supporting of this work.
References
- Ashraf G. Heterologous expression of stress-responsive DUF538 domain containing protein and its morpho-biochemical consequences. Plant J. 2011;30(5):351-8.
- Takahashi S, Yoshikawa M, Kamada A, et al. The photoconvertible water-soluble chlorophyll-binding protein of Chenopodium album is a member of DUF538, a superfamily that distributes in Embryophyta. J Plant Physiol. 2013;170(17):1549-52.
- Ashraf G, Kohnehrouz SB.Identification ofDUF538cDNA clone from Celosia cristata expressed sequences of nonstressed and stressed leaves. RussJPlant Physiol. 2010;57(2):247-52.
- Nakagami H, Sugiyama N, Mochida K, et al. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010;153(3):1161-74.
- Ashraf G, Kohnehrouz SB. ProtJ.2013;32:163.
- Ashraf G.Prediction of tertiary structure homology between bactericidal/permeability increasing protein of innate immune system and hydrolase enzymes. IntJBiosci. 2014;5(2):1-6.
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seiolman JG, Smith JA, et al. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1991.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402-8.
- Rabbani MA, Maruyama M, Abe H, et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant physiol. 2003;133(4):1755-67.
- Shinozaki K, Yamaguchi SK, Seki M, et al. Regulatory network of gene expression in the drought and cold stress responses.CurrOpinPlant Biol. 2003;6(5):410-7.
- Shinozaki K, Yamaguchi SK. Gene networks involved in drought stress response and tolerance. JExpBot. 2007;58(2):221-7.
- Yamaguchi SK, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10(2):88-94.
- Bartels D, Sunkars R. Drought and Salt Tolerance in Plants. CritRevPlant Sci. 2005;24(1):23-58.
- Seki M, Narusaka M, Abe H, et al. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13(1):61-72.
- Schimid M, Davison TS, Hens SR, et al. A gene expression map of Arabidopsis thaliana development. NatGenet. 2005;37(5):501-6.